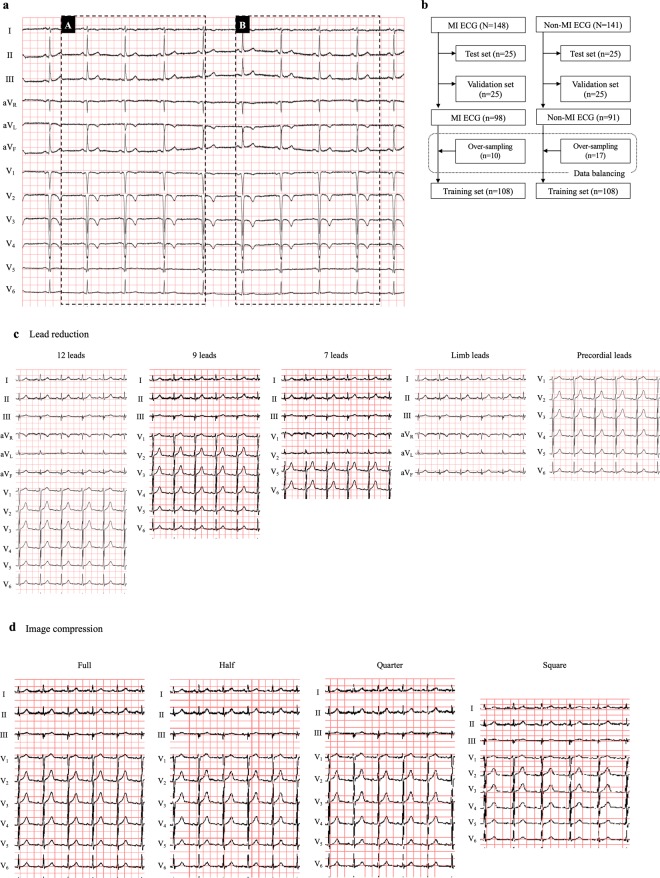
Figure 2.
ECG data and datasets preparation. (a) Target ECG regions for the present study are shown. The region A (744*368) was extracted for our deep learning processes. The region B (744*368) was used for over-sampling in order to resolve the data imbalance (see text in detail). (b) Dataset preparation is shown in this flowchart. As test sets and validation sets, 25 ECGs were randomly selected respectively. For data balancing, 10 or 17 ECGs were randomly selected in each group (over-sampling, see Fig. 2a), and added to construct the training sets. (c) ECGs with lead reduction are shown. 9-lead-ECG consisted of leads I, II, III, and V1–6. 7-lead-ECG consisted of leads I, II, III, V1–2, and V5–6. (d) Based on the 9-lead-ECG, we compressed its image quality in 4 different ratios; full quality (744*368), half quality (398*260), quarter quality (282*184) and square-shape (224*224).