Figure 1.
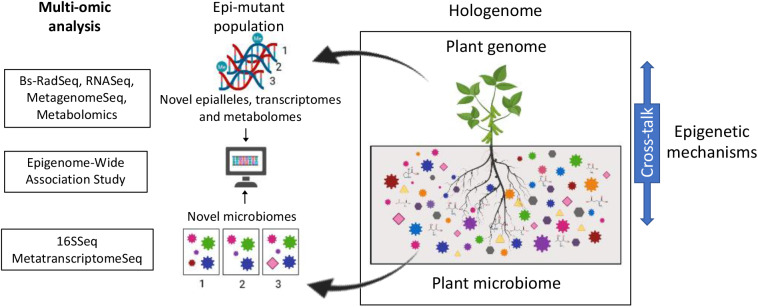
Proposed model for the identification of genes regulating plant-microbe interactions via the stochastic perturbation of DNA methylation patterns in plant populations. Exogenously induced DNA demethylation randomly generates novel epialleles in the plant population. These in turn alter the expression of genes which will alter the plant phenotype (i.e., microbiomes) of the plants carrying such novel alleles. Changes in DNA methylation in the plant population can be assessed using reduced representation approaches such as BsRADSeq. Changes in microbiome composition and functionality can be assessed using 16S sequencing and/or Meta-transcriptome analysis. Association between individual changes in DNA methylation (novel epialleles) and changes in phenotype (novel microbial community composition and functions) will be determined using epigenome wide association studies. Analysis of plant gene expression and metabolite production will be used as a validation step of the identified associations.