Figure 2.
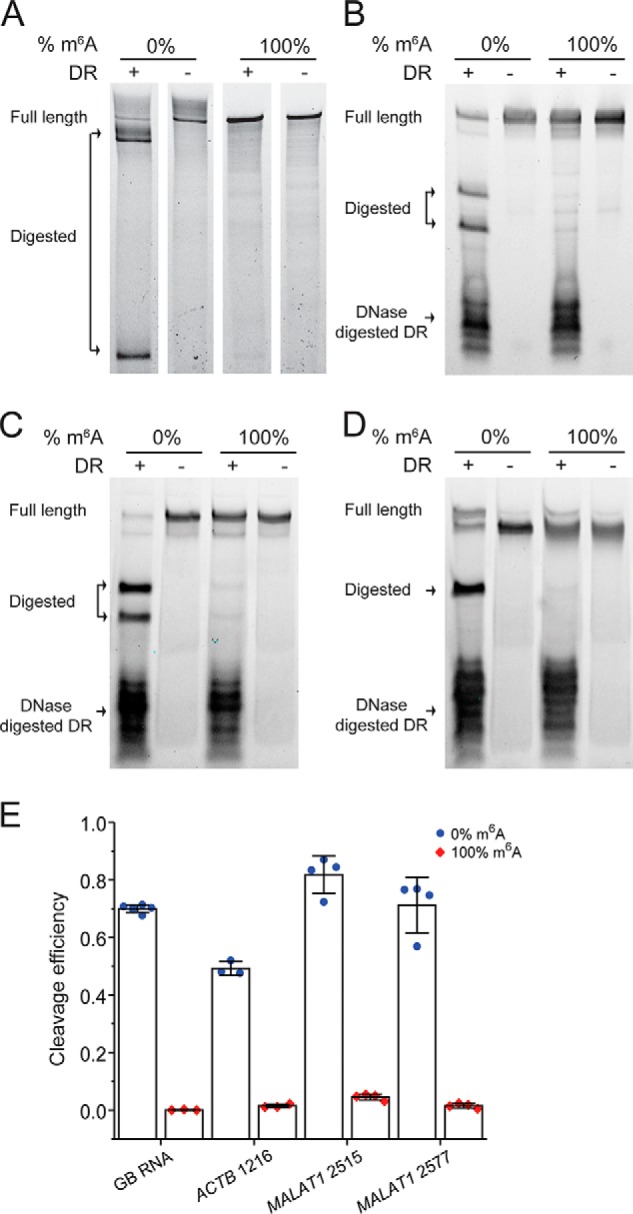
DR specifically cleaves unmodified RNAs, and its cleavage efficiency depends on sequence context around m6A sites. A–D, PAGE showing DR cleavage of 0 and 100% modified GB RNA (A) and RNA fragments containing modification site of ACTB 1216 (B), MALAT1 2515 (C), and MALAT1 2577 (D). E, bar plot of the cleavage efficiencies of m6A modified and unmodified target sites as quantified from PAGE. The error bars indicate means ± S.D. of three or four independent cleavage reactions. The DR is digested with DNase after DR cleavage reaction, because the DR may migrate to the same location as the input or cleaved RNA and affect data interpretation.
