Abstract
Myosin II is the main force-generating motor during muscle contraction. Myosin II exists as different isoforms that are involved in diverse physiological functions. One outstanding question is whether the myosin heavy chain (MHC) isoforms alone account for these distinct physiological properties. Unique sets of essential and regulatory light chains (RLCs) are known to assemble with specific MHCs, raising the intriguing possibility that light chains contribute to specialized myosin functions. Here, we asked whether different RLCs contribute to this functional diversification. To this end, we generated chimeric motors by reconstituting the MHC fast isoform (MyHC-IId) and slow isoform (MHC-I) with different light-chain variants. As a result of the RLC swapping, actin filament sliding velocity increased by ∼10-fold for the slow myosin and decreased by >3-fold for the fast myosin. Results from ensemble molecule solution kinetics and single-molecule optical trapping measurements provided in-depth insights into altered chemo-mechanical properties of the myosin motors that affect the sliding speed. Notably, we found that the mechanical output of both slow and fast myosins is sensitive to the RLC isoform. We therefore propose that RLCs are crucial for fine-tuning the myosin function.
Keywords: actin, myosin, molecular imaging, optical tweezers, single-molecule biophysics, actin filament gliding assay, myosin II, regulatory light chain, solution kinetics, optical trapping, single-molecule analysis, molecular motor, muscle contraction
Introduction
Myosin motors drive diverse motile processes, ranging from intracellular cargo transport and cell division to muscle contraction and the whole cell movement. Striated muscle myosin, such as skeletal or cardiac myosin II, responsible for generating the force during muscle contraction is a hexameric motor composed of two heavy chains (myosin heavy chains (MHCs)6) and four light chains (LCs) (1). Each heavy chain contains a globular motor domain, an α-helical light-chain binding domain, and a long coiled-coil rod. The rod parts from different myosin molecules self-assemble to form bipolar thick filaments. The ∼9-nm-long α-helix, also termed as the “lever arm,” serves as a link between the motor domain and the rod part of heavy chains. Each MHC has an essential (ELC) and regulatory light chain (RLC) wrapped around the lever arm (see Fig. 1A). The main motor domain has ATP- and actin-binding sites. During the chemo-mechanical coupling, small conformational change in the catalytic domain linked with the ATP hydrolysis gets amplified as a large movement of the lever arm (2, 3). The light chains are reported to be essential to maintain the rigidity of the lever arm as force is transduced during the power stroke (4).
Figure 1.
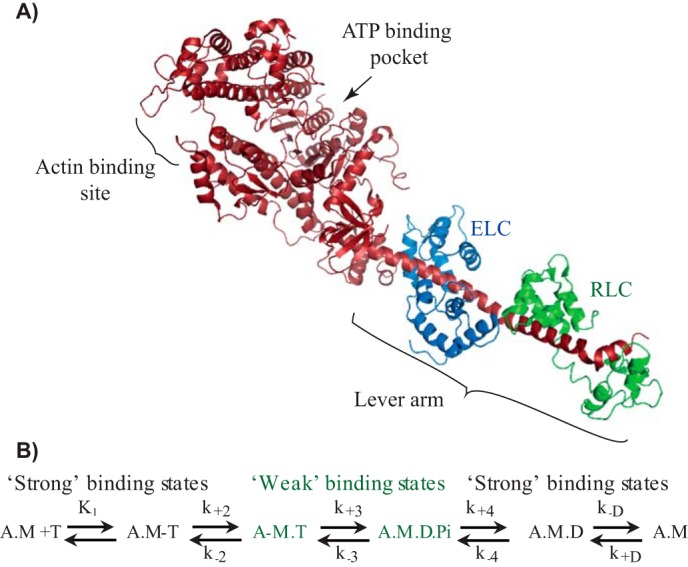
Myosin S1 structure and ATPase cycle. A, crystal structure of scallop myosin II-subfragment-1 (Protein Data Bank code 1SR6) (62). The myosin heavy and light chains are shown as a ribbon diagram; S1 is indicated with the key parts of the motor domain. B, kinetic scheme for actomyosin ATPase. Myosin II with different nucleotide states is illustrated: A, actin; M, myosin; T, ATP; D, ADP; Pi, Pi. Strong and weak interaction states of myosin with actin are labeled in black or green, respectively. Rate constants for forward and reverse reactions are indicated as k+n and k−n, respectively.
Muscle myosin II exists as different isoforms, three “fast” myosin heavy chain (MHC) isoforms (i.e. MHC-IIa, -IId/IIx, and -IIb) and one “slow” MHC-I isoform (5). The MHC-I isoform or β cardiac isoform is primarily expressed in slow skeletal, aerobic muscle fibers and in heart ventricle (6). The muscle expressing “fast” MHCs expresses myosin light-chain (MLC) isoforms (i.e. different ratios of ELC isoforms, MLC1f and MLC3f, and RLC isoform, MLC2f). The “slow” MHC-I isoform is typically equipped with the ELC isoform, MLC1s, and the RLC isoform, MLC2s/MLC2v.
RLC or MLC2 (∼19 kDa), encoded by the MYL2/Myl2 genes, is a member of the EF-hand superfamily of Ca2+-binding proteins with a helix-loop-helix structural motif. MLC2 noncovalently binds to the distal end of the lever arm at the junction between the S1 and S2 regions of myosin. Whereas the C-terminal domain of RLC binds in the region between Glu-808 and Val-826 of myosin lever arm, the N-terminal domain of RLC surrounds the region between amino acids Asn-825 and Leu-842 (2). Apart from its structural role to provide rigidity, RLCs are known to regulate cardiac, smooth, and skeletal muscle contraction via phosphorylation. For example, genetic loss-of-function studies in mice revealed an essential function of RLCs in cardiac contraction (7). Compelling reports demonstrated further that RLC phosphorylation regulates cardiac muscle contraction by increasing the number of cross-bridges available to generate the force (8–10). Reduction in phosphorylation levels of MLC2v are critically linked with human cardiomyopathies (11). Phosphorylation of MLC2 in smooth muscle myosin II determines the active state of the motor required for muscle contraction, thus making RLC a key regulator. However, for striated muscle myosin, the phosphorylation status of RLC is not correlated with the activation of the cross-bridge cycle, but rather Ca2+ binding to troponin activating the thin filaments is the prerequisite to initiate the muscle contraction. MLC2 phosphorylation is known to increase the steady-state force in permeabilized muscle fibers by increasing the number of cross-bridges interacting with actin filaments at submaximal levels of Ca2+ activation, without affecting maximum shortening velocity (12, 13). Thus, RLCs exert differential effects in a tissue-specific manner. Collectively, MLC-2 is thus considered to play a mechanical role by stabilizing the lever arm and a regulatory role through phosphorylation, but not to affect the ATPase kinetics.
Muscle fiber studies indicated that the cross-bridge kinetics underlying the force transients are determined mainly by the MHC isoforms (14). While the MHCs as the main determinant of the mechanical output is well-agreed on, MLC's primary role in skeletal myosin II is considered to be rather structural. The correlation of the muscle fiber–specific expression and assembly of the light chains with specific MHCs, however, might imply an additional role for the regulatory light chains. Two major observations—1) the distinct sets of light chains associating with different isoforms of myosin heavy chain and 2) single point mutations in the ventricular RLC causing heart disorder in humans, such as hypertrophic and dilated cardiomyopathies—directed us to investigate the importance of RLC in mechanical performance of the myosins.
This investigation is also relevant to understand the mechanical performance of the hybrid/chimeric motors (i.e. combination of specific myosin heavy chains with different light chains) found under different physiological conditions. In some cases, heterogeneity in MHC and LC isoform expression in single muscle fibers has been documented in vertebrate skeletal muscles. For instance, extensive analysis of the SDS-polyacrylamide gels from single soleus muscle fibers revealed that in a majority of fibers, the slow heavy chain and corresponding light chains were expressed. In some soleus single fibers, however, fast light-chain isoforms were expressed, which potentially may form a complex with the slow heavy chains (15). In human fast single fibers, co-existence of both fast (MLC2f) and slow (MLC2s) light chains was observed (16). Moreover, analyses of single fiber segments revealed coexpression of fast- and slow-myosin components nonuniformly distributed along the fiber length (17–19). During the developmental phase, the concentration of such hybrid/chimeric motors is reported to be increased (19). Thus, a mixture of fast and slow motor components (i.e. heavy and light chains) indeed exists in single muscle fibers; however, the mechanical properties of such hybrid motors remain unexplored.
Here, we examined the myosin chimeras where the endogenous regulatory light chain was replaced with a different RLC variant to probe whether, how, and which biochemical and mechanical properties are affected as a result of different trimeric formation(s) of myosin II motor complex (i.e. MHC, ELC, and RLC). We found that the motor properties of a given myosin isoform were sensitive to the type of RLCs. A chimeric motor based on fast myosin heavy chain translocated the actin filaments with reduced speed; conversely the slow myosin improved the gliding speed, simply by replacing native RLC with a different RLC variant. Steady-state and transient ATPase kinetics measurements allowed us to relate the altered motor properties to changes in the overall ATP turnover time and rates of product release. Single-molecule analyses based on optical trapping measurements revealed pronounced alterations in the duration of the strong actin-bound states, size of the power stroke, and stiffness of motors. These studies unraveled the critical role of different RLCs in muscle myosin motor function.
Results
Reconstitution of chimeric myosin II motors and analysis of actin filament gliding
We investigated the role of the RLCs in modulating the motor function of skeletal myosin II isoform found in the fast- and slow-twitch muscle fibers. We isolated myosin II motors from rabbit fast-twitch (musculus psoas) and slow-twitch (musculus soleus) muscle fibers. We inspected the native fast and slow myosin extracted from respective muscles in the SDS-polyacrylamide gels to ensure the purity (Fig. S1). We generated single-headed subfragment 1 (S1) and reconstituted the chimeric motors based on fast (MHC-IId) and slow (MHC-I) myosin II. In its native form, MHC-IId, henceforth referred as WT fast (WT-S1f) myosin S1, associates with the 2 light chains, i.e., ELC, MLC1f/MLC3f, and the RLC, MLC2B. MHC-I or WT slow (WT-S1s) myosin S1 is typically equipped with MLC1v and MLC2v.
As shown in Fig. 2A, we used three different RLCs (i.e. human fast skeletal regulatory light chain (MLC2B), human slow skeletal or ventricular RLC (MLC2v), and chicken gizzard smooth muscle myosin RLC (cgmRLC)) to generate six different chimeric motors based on the fast and slow myosins. The cgmRLC was previously reconstituted with the chicken skeletal myosin II and was shown to retain the high gliding velocity of the motor (20). For smooth muscle myosin (SMM) lower gliding speeds from 0.2 to ∼1 μm/s were reported under different motility conditions (21–23). SMM displayed kinetics properties similar to those of slow skeletal soleus myosin (6), with basal Mg2+ ATPase activity of 0.05 s−1, acto-S1 ATPase Vmax of 0.7 s−1, ADP dissociation of 15 s−1 (at 20 °C), and a second order rate constant for ATP binding of 0.5 μm−1 s−1 (24). Despite these SMM properties, when it was assembled with chicken skeletal muscle myosin, the actin filament gliding speed of the myosin subfragment 1 was increased by ∼4-fold, similar to the native full-length skeletal myosin (20).
Figure 2.
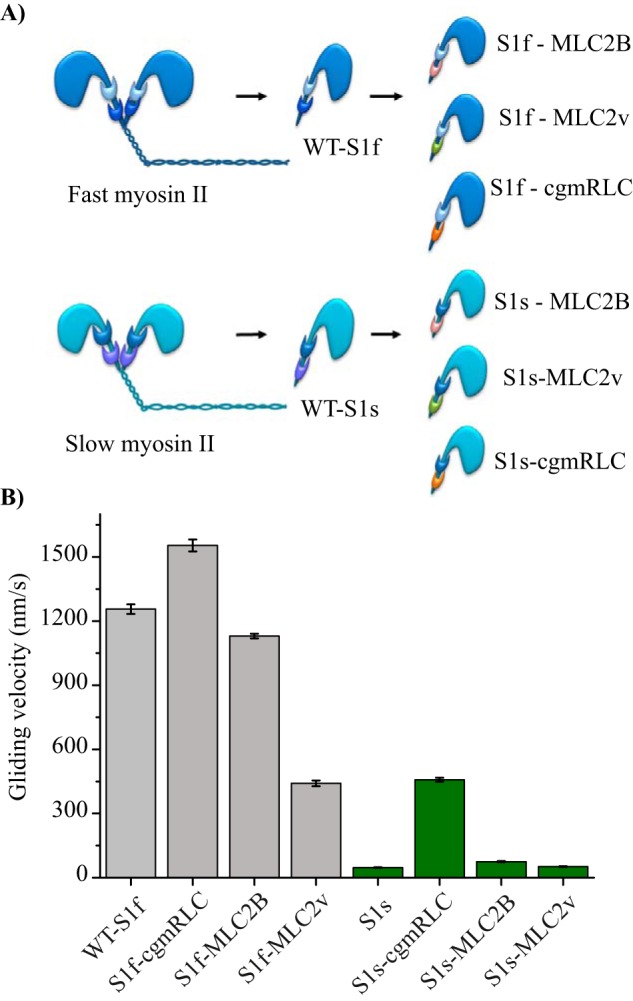
Reconstitution of chimeric motors. A, scheme illustrates in vitro reconstituted motors with different combinations of RLCs with fast (dark blue) and slow myosin II (sky blue) heavy chains. Papain digestion of full-length myosin II generated single-headed myosin S1. Regulatory light chains are color-coded to indicate the exchange: MLC2B (pink), MLC2v (green), and cgmRLC (orange). B, in vitro motility assay using native myosin S1 (WT-S1f and WT-S1s) or with chimeric motors. Motors were immobilized on a nitrocellulose-coated surface. Speed of movement was measured at saturating ATP concentration of 2 mm at room temperature (22 °C). Bar diagrams show the reduction in mean velocity from 1.2 ± 0.19 for WT-S1f to 0.44 ± 0.13 μm/s for S1f-MLC2v (N = 72 and 100 actin filaments, respectively) and the increase in mean velocity for WT-S1s from 0.047 ± 0.05 to 0.40 ± 0.05 μm/s for S1s-cgmRLC (N = 82 and 166 filaments, respectively). The motility experiments were performed with at least three different preparations of myosin motors and chimeras and were highly reproducible. For S1f-cgmRLC, N = 70; for S1f-MLC2B, N = 110; for S1s-MLC2v, N = 56; and for S1s-MLC2B, N = 55. Error bars, S.E. Statistical significance was calculated using unpaired t test for the following pairs of motors: WT-S1f and S1f-cgmRLC, p < 0.0001; WT-S1f and S1f-MLC2v, p < 0.0001; WT-S1s and S1s-cgmRLC, p < 0.0001; WT-S1s and S1s-MLC2B, p < 0.0001.
Therefore, we used it as a control to test its effect on the fast and slow skeletal myosin under our experimental conditions. As depicted in the Fig. 2A, we replaced the endogenous RLCs of WT myosins and generated six chimeric forms. The fast myosin chimeras comprised, 1) S1f-MLC2B (i.e., fast MHC with skeletal RLC), 2) S1f-MLC2v (i.e. fast MHC with ventricular RLC), and 3) S1f-cgmRLC (i.e. fast MHC with smooth mucle RLC). The slow myosin chimeras included, 1) S1s-MLC2v (i.e. slow MHC with ventricular RLC), 2) S1s-cgmRLC (i.e. slow MHC with smooth mucle RLC), and 3) S1s-MLC2B (i.e. slow MHC with skeletal RLC). We employed human variants of the RLCs because of the nearly identical protein sequences as shown in Fig. S2. Human MLC2B (MLC2B) and rabbit MLC2f (rMLC2f) share 98% sequence similarity. Likewise, human MLC2v is 98% similar to the rabbit slow RLC isoform. However, the fast isoforms MLC2B and rMLC2f has an 86% sequence similarity with the slow MLC2v. The cgmRLC shares low sequence similarity with both fast MLC2B and slow MLC2v (i.e. only 67%).
To understand the main effect of RLCs in these different combinations of chimeric motors, we focused on the mechanical parameters at the ensemble level using an in vitro actin filament gliding assay. We characterized two WT and six chimeric motors in in vitro actin filament gliding assays performed at 2 mm ATP concentrations (and 50–100 μg/ml surface density of motors). The motors were immobilized on a nitrocellulose-coated surface. As shown in Fig. 2B and Table 2, WT-S1f and WT-S1s showed >20-fold difference in the gliding speed (i.e. 1.25 and 0.047 μm/s, respectively) (see Movie S1). Very interestingly, we found gliding velocities on both fast MHC-IId and slow MHC-I motor surface to be sensitive to the variant of RLCs bound to the lever arm. The S1f-MLC2v combination resulted in a decrease in the actin filament gliding velocity by ∼3-fold, whereas S1f-cgmRLC and S1f-MLC2B retained the high actin filament gliding speed similar to the WT-S1f. On the contrary, the S1s-cgmRLC chimeric motor showed an ∼10-fold increase in the gliding speed of the slow S1s motor from 0.047 to 0.43 μm/s. The S1s-MLC2B motor showed a marginal increase in the gliding speed compared with WT S1s. Interestingly, as shown in Movie S2 and Fig. 2B, fast and slow myosin-driven actin filaments were indistinguishable when fast MHC was combined with MLC2v (S1f-MLC2v) and slow MHC with cgmRLC (S1s-cgmRLC).
Table 2.
Summary of ensemble molecule motility assay and parameters from single-molecule optical trapping measurements
Shown are actin filament gliding speed, lifetimes of AM-bound state, displacement of actin by individual motors, and stiffness for the WT and chimeric motor forms.
| Motors | Ensemble molecule analysis |
Single-molecule analysis |
||
|---|---|---|---|---|
| Gliding speed | Lifetimes | Displacement | Stiffness | |
| μm/s | ms | nm | pN/nm | |
| WT-S1f | 1.26 ± 0.022 | 46.4 ± 1.42 | 4.04 ± 0.41 | 1.12 ± 0.03 |
| S1f-cgmRLC | 1.56 ± 0.027 | 30.6 ± 7 | 3.99 ± 0.35 | |
| S1f-MLC2B | 1.13 ± 0.011 | 46.5 ± 2.6 | 5.63 ± 0.20 | 1.24 ± 0.09 |
| S1f-MLC2v | 0.441 ± 013 | 17 ± 0.4 | 4.34 ± 0.331 | 1 ± 0.07 |
| WT-S1s | 0.047 ± 0.001 | 282 ± 21.2 | 6.11 ± 0.16 | 0.437 ± 0.08 |
| S1s-cgmRLC | 0.458 ± 0.098 | 78.7 ± 8.2 | 6.21 ± 0.29 | 0.585 ± 0.024 |
| S1s-MLC2B | 0.075 ± 0.002 | 106 ± 9 | 1.89 ± 0.22 | 1.62 ± 0.27 |
| S1s-MLC2v | 0.052 ± 0.002 | 297 ± 11 | 4.25 ± 0.33 | 0.533 ± 0.11 |
These experiments showed that the motile activity of skeletal muscle myosin II is critically influenced by the regulatory light-chain isoform. Replacement of the native RLCs from fast and slow myosin II S1 with the RLC variants, MLC2B and MLC2v, respectively, showed comparable sliding velocities (Fig. 2B and Table 2). These results ruled out the alteration of motor properties due to the exchange procedure and further emphasized RLC-specific effects on myosin function.
The altered sliding velocities indicated that RLC interferes with the actomyosin (AM) chemo-mechanical coupling. Actin filament sliding velocities are governed by the net rate of actomyosin cross-bridge cycling (25, 26). The parameters that influence the sliding speed (V) include (i) the lifetime of the strong AM-bound state (ton) and (ii) the stroke size (d) of myosin (i.e. V = d/ton). The duration of AM-bound and -unbound states during the ATPase cycle described by duty ratio of the myosin is the fraction of the total ATPase cycle time myosin spends bound to actin. The duty ratio can change either by a change in the ADP release rate from the AM complex or the rate of weak-to-strong bound transition of AM (i.e. A.M. D.Pi to A.M.D state) (see the ATPase scheme in Fig. 1). One other parameter that must be considered in ensemble molecule sliding velocity measurements is the surface density of the myosin molecules. The skeletal myosin II, a low-duty ratio motor (<0.05), however, showed low dependence of velocities on the motor density (27, 28). Therefore, we presumed that the velocities most likely changed due to altered ton and/or d.
For native S1f, while some reports showed an isomerization step before Pi release as a rate-limiting step, the others suggested Pi release as the slowest step in the AM ATPase cycle (29, 30) and ADP release to be fast (>500 s−1). For the WT S1s, however, ADP was shown to have >10-fold higher affinity and thus slower ADP release during the cross-bridge cycle, ∼20 s−1 (6). Therefore, the detachment rate of the AM complex at the post-powerstroke state is limited by either the rate of ADP release or ATP binding to the rigor AM complex for these slow and fast myosin isoforms.
Ensemble molecule steady-state ATPase and transient kinetics
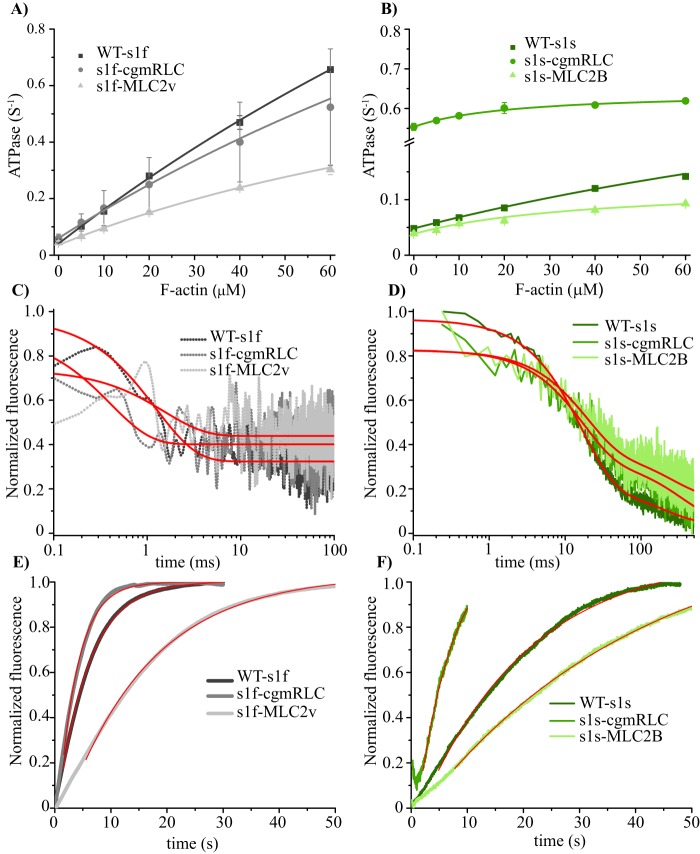
To investigate the cause of the altered sliding velocities, we measured actin-activated ATPase activities and rates of product release to determine potential rate-limiting steps in the ATPase cycle (Fig. 3). Consistent with previous studies (6), we obtained comparable basal ATPase activities between WT-S1f (0.04 s−1) and S1s (0.05 s−1) and almost 100-fold (3.5 s−1) and 10-fold (0.5 s−1) increase in the actin-activated ATPase activities, respectively (Fig. 3 (A and B) and Table 1). S1f-cgmRLC displayed comparable actin-activated ATPase activity as the WT construct (Table 1), whereas S1f-MLC2v showed reduced actin activation, as reported previously (31) (Table 1). For the slow myosin chimeras, we observed pronounced differences in the basal ATPase activity by almost a factor of 10 between S1s (0.05 s−1) and S1s-cgmRLC (0.55 s−1). The high basal ATPase activity of S1s-cgmRLC was unexpected but was reproducibly observed with different preparations. Such comparably high basal activities have been observed in mutant myosins (32, 33) and also unconventional myosins, myosin IX (34) and myosin VIIa (35). The S1s-MLC2B construct showed a similar basal activity as the WT S1s (0.04 s−1). The ATPase activity increased for all constructs with increasing actin concentrations, yielding values for activation at 60 μm F-actin of 0.14, 0.62, and 0.09 s−1 for S1s, S1s-cgmRLC, and S1s-MLC2B, respectively. Notably, the efficiency by which actin activated the ATPase (kcat/Kapp) was changed 3–5-fold among the motors (Table 1). In additional transient kinetic experiments, we measured the rates of ADP and Pi release as major determining factors of ATPase and sliding velocity. Generally, ADP release from actin-bound S1f is a very fast process with an estimated rate constant >500 s−1 (6). As seen in Fig. 3C, ADP-release kinetics was too fast to be resolved or not accompanied by signal change. The amplitude change was limited by the dead time (0.5 ms) of the instrument, providing only estimates for the rates of ADP release from actomyosin of >500 s−1 for all constructs (Table 1). Notably, ADP release from actin-bound S1s could be monitored due to its slower release kinetics and high ADP affinity (6). We observed two-step ADP-release kinetics for S1s, S1s-cgmRLC, and S1s-MLC2B, yielding rate constants of k′−AD = 54, 61, and 60 s−1 for the fast phase and k′−AD = 3.9, 4.9, and 2.9 s−1 for the slow phase, respectively (Fig. 3D). We observed Pi release as the rate-limiting step of the ATPase reaction for all measured constructs (Fig. 3, E and F). This is reflected in the similarity between Pi release rates and actin-activated ATPase rate at the 20 μm actin concentration (Fig. 3 (A and B) and Table 1). Thus, the RLCs appear to dictate the overall cycling time, mainly by affecting ADP release and modulating the Pi release kinetics.
Figure 3.
Ensemble kinetic experiments. Plots of ATP turnover rates as a function of F-actin concentration for fast (A) and slow (B) myosins. Data were fitted to hyperbolas using the Michaelis–Menten formula. The catalytic efficiency, kcat/Kapp, was obtained from the initial slope of the hyperbola. C and D, ADP-release kinetics from actomyosin. The rate of ADP dissociation from the actomyosin complex was measured by rapidly mixing actomyosin-mantADP complex with excess ADP. The decrease in mant fluorescence followed single exponential kinetics in the case of the fast myosins (C). For the slow myosin constructs, the fluorescence transients were best described by two exponentials with a fast rate constant (k−ADfast) and a slow rate constant (k−ADslow) indicative of two-step ADP-release kinetics (D). ATP turnover experiments of actomyosin constructs used coumarin-labeled phosphate-binding protein (MDCC-PBP; Thermo Fisher Scientific) as a sensor for the detection of liberated Pi from fast (E) and slow (F) myosins. The transients follow single exponentials with a corresponding rate constant for Pi release (k−Pi). All kinetic parameters are summarized in Table 1.
Table 1.
Summary of kinetic parameters
kbasal, ATPase activity in the absence of actin; vmax, maximum actin-activated ATPase activity; Kapp, concentration of actin at one-half vmax. vmax and Kapp were estimated from hyperbolic fits. vmax (60 μm), actin-activated ATPase rate at 60 μm F-actin; kcat/Kapp, catalytic efficiency; k−ADfast and k−ADslow, rate constants of the ADP release from acto-myosin; k−Pi, rate constant of Pi release at 20 μm F-actin concentration.
| Parameter | S1f | S1f cgmRLC | S1f MLC2v | S1s | S1s cgmRLC | S1s MLC2B |
|---|---|---|---|---|---|---|
| kbasal (s−1) | 0.04 ± 0.01 | 0.06 ± 0.01 | 0.04 ± 0.01 | 0.05 ± 0.01 | 0.55 ± 0.01 | 0.04 ± 0.01 |
| vmax (fit) (s−1) | 3.5 ± 1 | 2.5 ± 0.1 | 1.0 ± 0.3 | 0.5 ± 0.2 | 0.6 ± 0.1 | 0.1 ± 0.1 |
| vmax (60 μm) (s−1) | 0.66 ± 0.01 | 0.52 ± 0.2 | 0.30 ± 0.02 | 0.14 ± 0.01 | 0.62 ± 0.01 | 0.09 ± 0.01 |
| Kapp (fit) (μm) | 278 ± 96 | 244 ± 10 | 142 ± 54 | 208 ± 91 | 22 ± 3 | 39 ± 4 |
| kcat/Kapp (10−3 μm−1 s−1) | 10 | 11 | 4 | 2 | 4 | 2 |
| k−ADfast (s−1) | > 500 | > 500 | > 500 | 54 ± 3 | 61 ± 1 | 60 ± 3 |
| k−ADslow (s−1) | 4 ± 1 | 5 ± 1 | 3 ± 1 | |||
| k−Pi (20 μm) (s−1) | 0.18 ± 0.01 | 0.29 ± 0.01 | 0.07 ± 0.01 | 0.05 ± 0.01 | 0.17 ± 0.01 | 0.03 ± 0.01 |
Single-molecule analysis
To further probe the change in velocities with chimeric motors, we employed a single-molecule analysis method to gain precise insights into biochemical properties, such as the duration of AM strong bound states, and biophysical properties, such as stroke size (d) and stiffness of native and chimeric motor forms.
Single chimeric motor molecule's kinetic properties
We employed three-bead trapping assays for in-depth analysis of the biochemical and mechanical properties that led to either improved or decreased actin filament gliding speed driven by chimeric motors. Optical trap experimental setup has been described in detail previously (36). Briefly, the actin filament is embedded and stretched between the two optically trapped beads forming a dumbbell, and the motor is immobilized on a glass bead attached on the chamber surface (Fig. 4A). The positions of the two trapped beads are precisely monitored using a quadrant photodiode and acquired.
Figure 4.
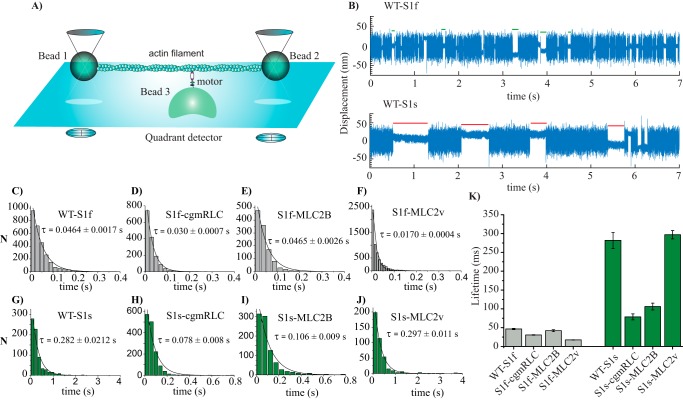
Single-molecule optical trapping: lifetime of AM interactions. A, the experimental setup for three-bead optical trapping measurements. Note that the indicated components, bead size, and protein dimensions are not to scale. B, original displacement over time data records; actin-myosin interaction can be observed as a reduction in the large thermal fluctuations from a bead at 10 μm ATP. The top and bottom panels show records collected from fast and slow WT myosin S1, respectively. Some of the interaction events are shown as green (fast WT) and red (slow WT) lines to indicate the duration of lifetimes, ton. Measurements were done by applying positive feedback and triangular wave (∼600 Hz). C–J, for WT-S1f, WT-S1s, and chimeras, event lifetimes (ton) are plotted in histograms and fitted with a single-exponential decay function to determine the average time constant. K, the bar diagram shows the average ton for WT and chimeric motors at 10 μm ATP concentration and room temperature of ∼22 °C. Error bars, S.E. (from the fits). For WT-S1f, n = 5000 events, N = 21; for WT-S1s; n = 1675 events, N = 19, for S1f-cgmRLC; n = 1517 events, N = 10, for S1f-MLC2B; n = 1953 events, N = 16, for S1f-MLC2v; n = 5714 events, N = 22, for S1s-cgmRLC; n = 1468 events, N = 12, for S1s-MLC2v; n = 400 events, N = 4, for S1s-MLC2B; n = 900 events, N = 8 (where N = number of individual motor molecules and n = number of AM interaction events). Statistical significance was calculated for the following pairs of motors using nonparametric Mann–Whitney U test: WT-S1f and WT-S1s, p < 0.00001; WT-S1s and S1s-cgmRLC, p < 0.0001; WT-S1s and S1s-MLC2B, p < 0.0001; WT-S1f and S1f-MLC2B, p = 0.0691; WT-S1f and S1f-MLC2v, p < 0.0001, WT-S1f and S1f-cgmRLC, p = 0.114; WT-S1s and S1s-MLC2v, p = 0.0941. p < 0.05 was considered statistically significant; p > 0.05 was considered not statistically significantly different.
Single-headed WT or chimeric motor molecules were analyzed for their intermittent interaction with the actin filaments as shown in Fig. 4B at 10 μm ATP concentration. We determined the lifetimes of the motor interaction with actin. The reduced Brownian noise in the data traces was characteristic of the AM-bound states, ton, whereas free dumbbell noise, toff, represented myosin-detached states. It is well-known that actin accelerates the Pi release, and Pi release from the myosin active site is closely associated with the powerstroke generation. However, there is no clear consensus on whether Pi is released before, during, or after the powerstroke. In our experimental set up, the initiation of actomyosin association seen as the reduction in the noise amplitude of the bead coincides with the powerstroke generation.
Thus, ton comprised the post-powerstroke states with ADP present in the active site (AM.ADP) plus the time the active site is free of nucleotide (rigor state) until a new ATP molecule binds and initiates the rapid detachment of the myosin head from the actin filament. This argument is valid if the Pi is released before or during the powerstroke. Some recent studies have supported the model that the powerstroke precedes the Pi release from the active site (37, 38). However, to consider the AM.ADP.Pi state as a “strong bound state”—contributing to the lifetime of an attached event—the duration of this state should be sufficiently long after the powerstroke (at least 2 ms (i.e. the detection limit with our current experimental setup)). We cannot distinguish between strong bound AM.ADP.Pi and AM.ADP states.
The AM-detached state, toff, included the ATP hydrolysis and M.ADP.Pi state prior to association with actin (see the ATPase scheme in Fig. 1). As the ADP release from the active site is in the range of >500 s−1, we believe that for the fast myosins, primarily an AM-bound rigor state is acquired. At saturating ATP concentrations, the duration of the rigor state becomes too short and is essentially undetectable for fast myosins. Therefore, for comparison among different native and chimeric motors, we measured lifetimes, ton, at 10 μm ATP concentrations.
Fig. 4B shows the comparison of 7-s traces obtained with WT-S1f and WT-S1s, containing several distinct single AM-binding events at 10 μm ATP. The obvious difference in the two traces with the two types of motors was the short versus long lifetime of AM-bound events. Consistent with previous observations, S1f showed shorter ton as compared with S1s (39). We estimated the average lifetime of AM-bound states by fitting the ton events from several molecules with a single exponential decay function (Fig. 4, C–J). Fig. 4K and Table 2 display the average lifetimes for WT and the chimeric motors. Duration of AM bound states for WT-S1f, S1f-cgmRLC, and S1f-MLC2B showed no significant difference. However, the S1f-MLC2v motor showed a >2-fold decrease in ton. For slow myosin, compared with WT-S1s, S1s-MLC2v retained the extended ton. Interestingly, with S1s-cgmRLC, the ton was significantly reduced by ∼4-fold, and by ∼3-fold for S1s-MLC2B. From the reciprocal of average time constants, we estimated the AM detachment rate constants and thereby the second-order rate constants of ATP binding (kATP) for fast myosins, kATP = 2.2 μm−1 s−1 for WT-S1f, 3.26 μm−1 s−1 for S1f-cgmRLC, 2.2 μm−1 s−1 for S1f-MLC2B, and 5.9 μm−1 s−1 for S1f-MLC2v. Our estimation of kATP for WT-S1f is consistent with previous studies (40, 41).
For all of the chimeric constructs except for one (i.e. S1f-MLC2v), the increase in the duration of the AM-bound state was associated with the decrease in the actin filament sliding velocity. The chimeric motors, S1f-cgmRLC and S1f-MLC2B, showing ton (or kATP) comparable with WT-S1f, also displayed similar gliding velocities (see Fig. 2B). A similar tendency was observed for WT-S1s and S1s-MLC2v.
Mechanical properties
Stroke size
Previously, single-molecule analysis of motors showed that the lever arm length affects the amplitude of displacement (i.e. stroke size, d) (42). As an accessory protein associated with the lever arm, whether light chain influenced stroke size was not explored.
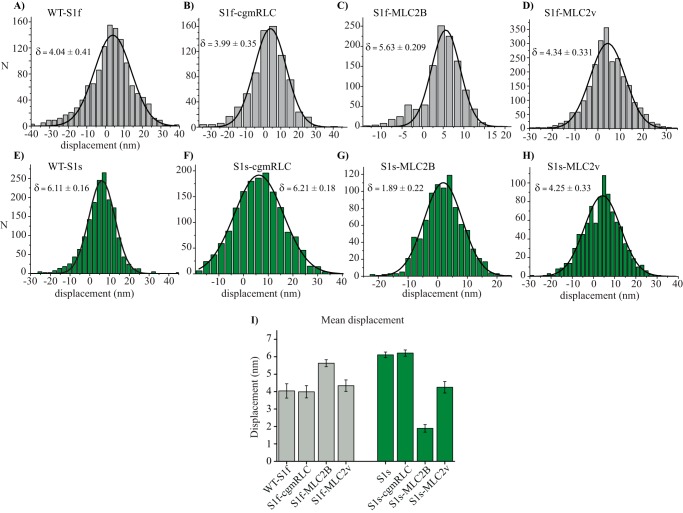
We therefore investigated whether different variants of the light chains influenced the amplitude of stroke size of the chimeric motors. We determined the average stroke size from the shift of histogram method, introduced by Molloy et al. (43). As shown in Fig. 5 (A–I) and Table 2, we noted a significant difference in the mean displacement for fast and slow motors (i.e. from 4.04 ± 0.41 to 6.11 ± 0.16 nm, respectively). Thus, consistent with previous reports for rat fast and slow skeletal myosin II-S1 (39), the average working stroke for WT-S1f was smaller than for WT-S1s. We found a moderate yet significant increase in the stroke size for chimeric construct S1f-MLC2B as compared with its WT counterpart. For slow myosin heavy chain, however, the S1s-MLC2v and S1s-MLC2B combination resulted in a significant decrease of the average displacement amplitude, with S1s-MLC2B displaying merely 1.86-nm stroke size.
Figure 5.
Powerstroke size of chimeric motors. A–H, the average stroke size/mean displacement was determined by histogram shift (δ) from mean free dumbbell noise. The histograms fitted with Gaussian function to determine the average stroke size for the indicated myosin motors. I, the average stroke size determined from the Gaussian fits for WT and chimeric motors were compared in a bar diagram. Error bars, S.E. from the fits. The statistically significant difference in the powerstroke size was calculated for the following pairs of motors: WT-S1f and WT-S1s, p < 0.0001; WT-S1f and S1f-cgmRLC, p = 0.8841; S1f and S1f-MLC2B, p = 0.0053; S1f and S1f-MLC2v, p = 0.0025; WT-S1s and S1s-cgmRLC, p = 0.1002; WT-S1s and S1s-MLC2B, p < 0.0001; WT-S1s and S1s-MLC2v, p < 0.0001. Statistical significance was calculated using unpaired t test.
Stiffness
The light chains are reported to be responsible for the structural stability of the lever arm. The ∼9-nm-long helical structure of lever arm that links the main motor core to the dimerization region and continues as a coiled-coil region that forms the basis of the thick filament backbone may contribute to the compliance of the motor.
One assumption is that variants of RLCs provide different stiffness to the myosin head. Consequently, different myosin head stiffness may result in a different mechanical strain. The kinetics of some steps of the ATPase cross-bridge cycle is assumed to respond to the change of strain under load (44, 45). The strain dependence of ATPase kinetics is well-reported for the motors, such as myosin V and smooth muscle myosin (24, 46). Could the binding of MLC2v compared with MLC2B change, for example, the stiffness of myosin S1f and resultantly alter the AM cross-bridge cycle?
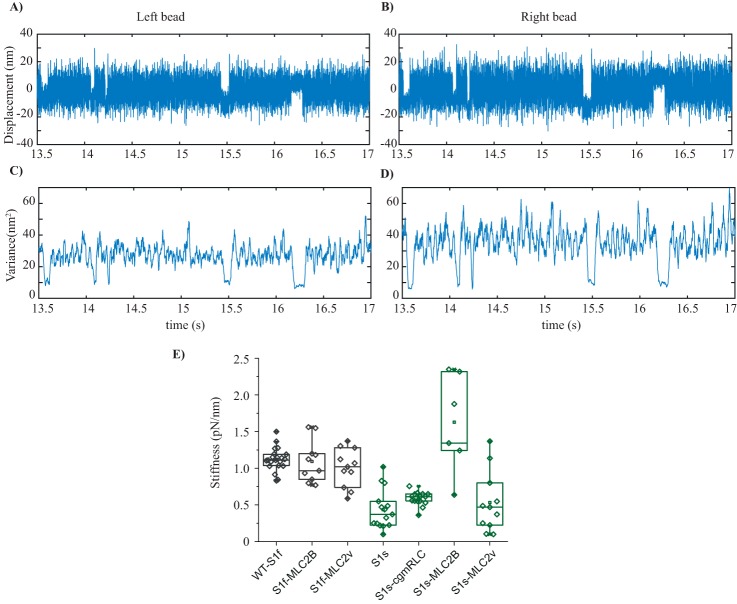
To investigate this notion of whether the different RLCs influenced the stiffness of the chimeric motors, we acquired the AM interaction records by applying positive feedback on the laser-trapped beads as described by Steffen et al. (36, 47). Accordingly, the signal/noise ratio (bound versus free dumbbell) was improved by increasing the noise amplitude of the free dumbbell, using analog positive feedback in AC mode. The trapped bead position analog signal from the quadrant detector was directed to the electro-optic deflectors, controlling the trap position. As the actomyosin binding event detection is more difficult for the motors, especially with low stiffness, this procedure allows the event detections and stiffness measurements by increasing the amplitude of Brownian noise by ∼2-fold, while maintaining the trap stiffness in the y and z directions. Using a bead variance-covariance method as described (48, 49), the stiffness of individual motors was calculated, as shown in Fig. 6. We found significant difference in the stiffness for WT-S1f versus WT-S1s (i.e. 1.12 ± 0.03 and 0.437 ± 0.08 pN/nm, respectively) (Fig. 6E). The stiffness values are comparable with previous optical trapping measurements from rat fast and slow myosin S1 (39, 50) and single fast and slow muscle fiber measurements for rabbit myosins (50). For the fast myosin, replacing the light chains with two different variants (MLC2B and MLC2v) did not alter the stiffness significantly, as shown in Fig. 6 and Table 2. Similarly, for slow myosin chimeras (S1s-cgmRLC and S1s- MLC2v), there was slight increase in the average stiffness, however not significantly different from the WT form. The S1s-MLC2B motor, however, showed the highest stiffness of 1.62 ± 0.27 pN/nm.
Figure 6.
Stiffness. A and B, original data records acquired in optical trapping experiments. Bead position is plotted over time for both the left and right bead of the dumbbell. Positive position feedback was used to increase the amplitude of thermal fluctuations, which effectively increased the variance ratio between binding events and free dumbbell noise for both traps in the direction of the actin filament axis. C and D, variance versus time plotted for the records shown in A and B. Variance was calculated for a rolling window of 20 ms, at our sampling rate of 10 kHz. For the example shown here, the variance-Hidden-Markov method yielded a combined trap stiffness of 0.078 pN/nm and a myosin head stiffness of 0.85 pN/nm. E, box plot with overlapped data points, the stiffness measured for WT-S1f, WT-S1s, and chimeric motors. Each rhombus represents the measured stiffness from individual myosins. Average stiffness with S.D. was as follows: for WT-S1f, −1.12 ± 0.03; for S1f-MLC2B, −1.24 ± 0.09; for S1f-MLC2v, −1 ± 0.07; for WT-S1s, −0.437 ± 0.08; for S1s-cgmRLC, −0.585 ± 0.024; for S1s-MLC2B, −1.62 ± 0.27; for S1s-MLC2v, −0.533 ± 0.11. Unpaired t test was used to calculate the statistical significance. WT-S1f (n = 20) and WT-S1s (n = 13) displayed statistically significant difference with p < 0.0001. No statistically significant difference between WT-S1f and S1f-MLC2B (N = 12, p = 0.72) or WT-S1f and S1f-MLC2v (N = 15, p = 0.128) was found. For WT-S1s and S1s-cgmRLC (N = 10), marginal but statistically significant difference was found (p = 0.0493). For WT-S1s and S1s-MLC2B, N = 6 and p < 0.0001. For WT-S1s and S1s-MLC2v, n = 11 and p = 0.4735. N = number of individual motor molecules.
To measure the stiffness of the WT-S1s and S1s-MLC2v chimera, the experiments were performed at higher ATP concentration (i.e. at 50 μm ATP) to avoid the occasional inclusion of contaminant fast myosin in the measurements. Although the soleus muscle primarily contains (about 95%) slow myosin II isoform, the presence of ∼5% fast myosin isoform cannot be avoided. In ensemble molecule gliding assays, 5% contamination of motors is unlikely to influence the actin filament speed as measured previously for mixtures of different ratios of myosin isoforms (22, 23). Accordingly, more than 20% contamination of motors was required to cause alteration of gliding speed. In the single-molecule assays, however, this factor needs to be cautiously controlled. At 50 μm ATP concentration, the AM interaction events for the fast myosin are too short for detection with our setup (<2-ms AM lifetime events could not be observed). This way we excluded the measurements from contaminant fast myosins. For S1s-cgmRLC and S1sMLC2B, stiffness was measured at 10 μm ATP concentration, as these motors showed ∼3-fold shorter ton as compared with the WT-S1s and S1s-MLC2v, and ∼2-fold longer average ton compared with the S1f chimeras.
Altogether, the mechanics measurement suggests that the light chains may cause substantial alteration of the amplitude of powerstroke size and the motor stiffness as seen for S1s-MLC2B. For the other chimeras, the parameters were either moderately altered or comparable with the WT forms (Table 2).
Discussion
For over 2 decades, the issue of myosin heterogeneity in the muscle fiber has been discussed in the literature, and particular emphasis has been put on the mixed content of myosin isoforms. However, until now, it was difficult to systematically examine the mechanical performance of myosins associating with different types of RLCs. Here, by reconstituting a homogeneous population of such well-defined hybrid/chimeric motors and analyzing the kinetic and mechanical features of the constructs using single-molecule and ensemble measurements, we were able to unravel the influence of individual RLCs on the mechanical performance of slow- and fast-muscle myosins.
By swapping the RLCs between fast- and slow-myosin heavy-chain isoforms, we showed that MLC2 naturally associating with fast myosin motor and smooth muscle myosin enhanced the originally “slow” motor's actin filament velocity, whereas the MLC2 that naturally assembles with the slow soleus or β-cardiac myosin slowed down the native fast myosin. It appeared that these specific MLC2 variants can increase or decrease the velocity, depending on the myosin heavy-chain isoform they are attached to. The changes in different kinetic and mechanical parameters of chimeric motors relative to the WT motor forms are listed in Table 1 and 2. In all but two chimeric motors (i.e. S1s-MLC2B and S1f-MLC2v), the ton that represents the duration of strong bound states correlated with the increase or decrease in the gliding velocity (Table 1 and 2). Although the trapping measurements were done at subsaturating ATP concentration, the relative changes in the ton indicated changes in AM detachment rates among WT and chimeric motors. Compared with its native forms, for chimeric motors, shorter and longer ton relating to higher and lower rates of actomyosin dissociation (1/ton), consequently resulted in faster or slower actin filament speed, respectively. The powerstroke size was comparable between WT-S1f and S1f chimeras and for WT-S1s and S1s chimeras, except for S1s-MLCB. S1s-MLCB displayed ton similar to that of S1s-CgmRLC, which displayed >5-fold higher gliding speed. However, the other differences between the two chimeras included a 3-fold shorter powerstroke size and slower rate of weak-strong transition (due to slow Pi release; Fig. 3F and Table 1) that may be responsible for the slower gliding speed for S1s-MLCB. Light chains are known to provide the rigidity to the myosin lever arm. It appears that this RLC isoform (MLC2B), when combined with the slow myosin heavy chain (S1s), influences the measured stiffness of the actomyosin cross-bridge. The decrease in the displacement (δ) of S1s-MLC2B is intriguing. One possibility could be that the association of the MLC2B hampers the complete swing of the lever arm to produce the displacement comparable with that of the native motors.
Typically, ADP release (strong bound state) rates are known to determine the gliding velocity for slow myosin. The solution kinetics measurement showed two phases of ADP release with marginal differences in the slow phase of ADP release rates for all slow myosin chimeras (Table 1). However, significant difference (up to ∼3-fold) in the ton was observed in trapping kinetics measurements under subsaturating ATP concentration, and the gliding speeds varied by up to ∼10-fold among WT and chimeric motors (S1s versus S1s-cgmRLC). A main difference between solution kinetics and the single-molecule trapping experiment is the load experienced by the motor. Thus, the observed differences in the parameters derived from the two methods could be explained by high sensitivity of the motors to the load as in trapping assays (even at the low applied values) and, consequently, load-sensitive ADP release influencing the mechanical output of the motors. In the solution kinetics measurement, the individual myosin molecules remain unaffected by the binding or release of other myosin heads. However, in the motility assay, several myosin molecules working as an ensemble to drive the actin filament movement are not completely independent but are mechanically linked through the actin filament (i.e. the motors may experience low levels of load).
For the chimera, S1f-MLC2v, the reduction in actin filament gliding speed could be associated with the increase in binding affinity to actin, a higher affinity for ADP, and slower ADP release rate, as shown previously (31). These changes may influence the actin-bound fraction of motors at any given time in the ensemble molecule gliding assay. If the bound motors are not coordinated, due to slower ADP release rates, the increased number of attached versus detached motors may generate a drag on actin filaments as compared with the WT counterpart. This may reduce the gliding velocity in the ensemble molecule assay. Consistent with increased duration of the actomyosin strong-binding state or a higher duty ratio for the S1f-MLC2v motor, we found that the dimeric chimera of fast heavy meromyosin, HMMf-MLC2v, even displayed moderately processive behavior along actin filaments in single-molecule studies (31). Remarkably, under single-molecule processive conditions, the HMMf-MLC2v chimera exhibited a high speed of movement (∼2.5 μm/s).
We identified a previously unrecognized aspect of the RLC's task (i.e. the effect on AM cross-bridge cycling by influencing the ATPase kinetics). An obvious question raised by our findings is how diverse RLCs attribute such a response on myosin function. This is a very intriguing yet quite complex question, which would require structural investigations and perhaps additional computational approaches to dissect possible long-range allosteric communication between the RLC and motor domain.
The different isoforms of RLC are similar in structure, as predicted from their aa sequences, in particular in the divalent cation-binding sites and proximal phosphorylatable serines in striated muscles (10). They have relatively low sequence similarity, with the light chains MLC2B/MLC2f and MLC2v 86% similar and the cgmRLC exhibiting only 67% sequence similarity with the fast MLC2B/MLC2f isoforms.
RLC is categorized as an EF-hand superfamily member, harboring helix-loop-helix structure and a cation (Ca2+ or Mg2+) binding site at the N terminus. Fig. S2 depicts the conserved eight predicted helices, which form four EF-hand domains. The binding of Ca2+ to the RLC is reported to induce a conformational change to a more open state whereby the overall helical content of the molecule increases, which subsequently positively affects the cross-bridge kinetics (51). In our measurements, we have no Ca2+ but have Mg2+ in the reaction buffers. Additionally, the aa sequences for the helix 1 and 2 regions are 100% identical in fast and slow RLC, whereas the loop that serves as the binding site for divalent cation has a 3-aa difference. With the current knowledge, however, it is challenging to foresee which specific feature(s) might define the particular modulating characteristics of RLCs. Detailed structural and functional investigations would be required to address the diverse impact of the regulatory light chains.
Our findings elucidate the specificity of RLCs toward distinct MHCs. In addition to understanding the fundamental role, precise details of the modulatory role of the RLC isoform are of particular clinical relevance, as single point mutations in the human myosin heavy (β-MHC) and regulatory light chains (MLC2v) have been linked to causing familial hypertrophic or dilated cardiomyopathy. Researchers have gained a plethora of valuable information not only from studies on human muscle tissue, but also from various animal models (e.g. mice and rabbits) and recombinant motor proteins about the motor dysfunction as a result of single point mutations. These studies often constituted motor components either from mixed isoforms (e.g. human mutant MLC2 with mice MHC and MLC1) or the mutations generated in different MHC isoform background (e.g. mouse α-MHC or Dictyostelium discoideum myosin II). Some studies were performed without light-chain binding domains or motors with only MLC1 and not MLC2.
Recent micro-mechanical characterization of human induced-pluripotent stem cell–derived cardiomyocyte myofibrils showed different (increased rates) kinetics of active force generation and relaxation from that of the adult human ventricular myofibrils. One possible explanation was that besides other protein isoform differences, the β-MHC expression did not fully match with the respective light-chain expressions (i.e. MLC1v and MLC2v); rather, atrial (MLC1a and MLC2a) light chains dominated over ventricle-specific light chains (52). Altogether, a striking revelation from our studies is the point that the complete “holoenzyme” with the respective heavy and light chains is critical to understanding the motor function and therefore should be taken into account in structure-function analyses.
Overall, our findings offer highly compelling evidence of the role of RLCs in modulating myosin motor function by influencing the actomyosin ATPase kinetics. The functional differences among the myosin II family members appear to be only partly defined by the myosin heavy chain alone. Here, we show that variations in light-chain composition crucially determine the mechanical output of a specific myosin II isoform that is commonly found in different muscle types and various tissues. The importance of these findings extends beyond basic understanding of the myosin function. The study opens new perspectives to investigate the physiological relevance of light chain–mediated fine-tuning of myosin function in the context of muscle contraction and additionally for other classes of myosin involved in transport processes.
Experimental procedures
Generation of single-headed myosin motors from native myosin II
Full-length rabbit fast skeletal muscle myosin II (MHC-IId) and MHC-I were isolated from skinned fibers of musculus psoas and musculus soleus muscle, as described previously (53, 54). The myosin II was digested with papain to generate single-headed subfragment 1 (myosin S1), respectively, as described earlier (55, 56). S1 was stored at −80 °C in 50% glycerol in a buffer (5 mm sodium phosphate, 10 mm potassium acetate, 4 mm magnesium acetate, and 2 mm DTT at pH 7.0). In the reconstitution experiments, freshly prepared S1 was employed for further downstream processing for light-chain exchange.
To collect the muscle tissues, the rabbits were euthanized as per the guidelines from German animal protection act §4 (Tötung zu wissenschaftlichen Zwecken/sacrifice for scientific purposes). The animal was registered under reference number 2016/122, obtained from Medical School Hannover central animal facility. All of the procedures were carried out in accordance with relevant guidelines and regulations from Hannover Medical School. No experiments were performed on live animals prior to the sacrifice.
Regulatory light-chain expression
The plasmid vector (EX-T0572-B09) containing full-length human cardiac, slow light-chain 2 insert (i.e. MYL2v) and vector (EX-D0356-B09) containing human, fast-skeletal muscle myosin regulatory light-chain (MYLPF) sequence were purchased from Genecoepia (Rockville, MD).The plasmid vector containing biotin-dependent transcarboxylase (BDTC) cgmRLC was kindly provided by Dr. Atsuko Iwane to Walter Steffen. The details of BDTC-cgmRLC fusion protein are provided by Iwane et al. (20). As mentioned by Iwane et al., a recombinant fusion protein, BDTC-cgmRLC was generated by fusing BDTC with chicken gizzard muscle myosin RLC sequence at the N terminus. BDTC is a 123-amino acid peptide sequence of the 1.3S subunit of Propionibacterium shermanii transcarboxylase. BDTC was post-translationally biotinylated by biotin holoenzyme ligase in Escherichia coli in vivo. Biotin was added to the bacterial culture in growth medium after the induction of the bacterial cells with isopropyl β-d-1-thiogalactopyranoside. The molecular weight of the BDTC-cgmRLC protein is ∼34 kDa.
The plasmids (EX-T0572-B09 and EX-D0356-B09) were transformed in RosettaTM competent cells (EMD Millipore, Burlington, MA) for expression of the light chains, MLC2v and MLC2B, respectively. The isolation and purification of the protein were as described previously (20, 56). Note that bacterially expressed regulatory light chains, BDTC-cgmRLC, MLC2B (or MLC2F), and MLC2v, were without post-translational modifications, such as phosphorylation.
Regulatory light-chain exchange
RLC exchange with S1 was performed as described previously (20, 31, 56–58), with some modifications. Myosins, S1f (psoas fast myosin subfragment 1) or S1s (soleus slow myosin subfragment 1), were incubated with an ∼10-fold molar excess of MLC2v/MLC2B/cgmRLC for 30 min at 30 °C in a buffer (Mg2+-free condition) containing 50 mm HEPES (pH 7.6), 0.5 m NaCl, 10 mm EDTA, 10 mm DTT. The exchange reaction was stopped by the addition of 12 mm Mg2+ to the reaction mixture and incubated on ice for 30 min. To remove the excess RLCs from the reconstituted myosin and to obtain the active motors, myosin was further incubated with high concentration of actin filaments for 30 min on ice. The mixture was centrifuged over 20% sucrose cushion buffer at 70,000 rpm for 30 min. The pellet was once washed with buffer (5 mm sodium phosphate, 10 mm sodium acetate, and 4 mm magnesium acetate, 2 mm DTT, pH 7.0) and then resuspended in the buffer with added 10 mm ATP to release active myosin from actin filaments. The actin filaments were separated from myosin solution by centrifugation. The supernatant containing myosin motors was mixed with 50% glycerol, flash-frozen in liquid nitrogen, and stored at −80 °C and used for further experiments as required. The probes were run on the 15% polyacrylamide gels and stained with Coomassie stain (Phastgel Blue R-350 from GE Healthcare). The gel images were acquired, and either ImageQuant or the ImageJ program was used for the densitometric analysis to determine the RLC exchange efficiency for the chimeric myosins. To measure the intensities of the RLC bands, the region of interest (including both the native and exchanged RLC bands) was selected and plotted as an intensity histogram. The area under the curve for each respective band was measured for the intensity values. The difference in the molecular weight of the protein was taken into account while calculating the exchange efficiency. About ≥85% RLC exchange was observed for several preparations of S1 as shown in Fig. S1. The myosin was used for motility and single-molecule assays. For the solution kinetics experiments, the myosin chimeras were prepared without the final actin co-sedimentation step to avoid ATP in the myosin solution. Instead, after the RLC exchange reaction was completed, the myosin was loaded on spin columns with high-molecular mass cut-off (100 kDa) to separate the free RLCs from the assembled myosin complexes. The removal of free RLC was ensured by multiple dilutions with buffer and passage through the spin columns. The myosin in buffer (5 mm sodium phosphate, 10 mm sodium acetate, 4 mm magnesium acetate, 2 mm DTT, pH 7.0) containing 3% sucrose was finally flash-frozen and stored at −80 °C.
Preparation of actin filaments
Actin filaments were generated by incubating rabbit G-actin in polymerization buffer containing 5 mm sodium phosphate, 10 mm sodium acetate, and 4 mm magnesium acetate, supplemented with protease inhibitor overnight at 4 °C. An equimolar concentration of fluorescent phalloidin was added to fluorescently mark the actin filaments. To obtain long biotinylated actin filaments (≥20 μm) for optical trapping experiments, additionally, 1 mm DTT and 1 mm ATP was added to the polymerization mixture as described by Steffen et al. (36).
Active myosin heads
Prior to use in in vitro motility assays or optical trapping measurements, the myosin was further purified to discard any inactive motors by co-sedimentation with concentrated F-actin. Actin-myosin complex was dissociated by the addition of 4 mm Mg·ATP. Whereas the active heads released from actin in the presence of ATP, the inactive myosin remained bound and separated by centrifugation at 70,000 rpm for 30 min at 4 °C. The supernatant containing the enzymatically active motors was further supplemented with 2 mm DTT and protease inhibitor and used in the functional assays. This procedure to remove the inactive motor heads was routinely followed prior to the main experiments.
Steady-state ATPase and transient kinetic experiments
Actin-activated ATPase was measured with a BioTek Synergy 4 multiplate reader (BioTek) using the NADH coupled assay. A solution of myosin (0.2 μm), F-actin, NADH (0.4 mm), lactate dehydrogenase (0.02 mg/ml), pyruvate kinase (0.05 mg/ml), and phosphoenolpyruvate (0.5 mm) in buffer containing 25 mm HEPES, pH 7.3, 25 mm KCl, 5 mm MgCl2, 0.5 mm DTT was mixed with ATP (2 mm) to start the reaction. ATPase rates at different concentrations of F-actin were obtained from the slopes of the corresponding linear fits of the time-dependent absorbance change at 340 nm. Rates were plotted against the F-actin concentration, and data were fitted according to Michaelis–Menten kinetics. Transient kinetic experiments were carried out with a Hi-Tech Scientific SF-61 DX double mixing stopped-flow system (0.5-ms dead time) at 20 °C. Experimental buffer contained 20 mm MOPS, pH 7.0, 25 mm KCl, 1 mm DTT, 5 mm MgCl2. For ADP-release measurements, a solution of 2 μm AM preincubated with 20 μm mantADP was rapidly mixed with 1.6 mm ADP. The reaction was followed through a KV389 cutoff filter by monitoring mantADP fluorescence changes induced by FRET through tryptophan excitation at 296 nm. Pi release was measured under single- or multiple-turnover conditions with ATP as substrate as described before (59).
In vitro motility assay
An in vitro motility assay was performed with monomeric S1 motors with native or exchanged RLC, by immobilization of the motors on a nitrocellulose-coated surface. The assay was described previously in more detail (53). Briefly, myosin was incubated for 5 min on a nitrocellulose-coated surface, followed by wash and surface blocking with 1 mg/ml BSA in assay buffer (25 mm imidazole hydrochloride, pH 7.2, 25 mm NaCl, 4 mm MgCl2, 1 mm EGTA, and 2 mm DTT). To block the inactive or damaged myosin motor heads, 0.25 μm short, unlabeled F-actin was injected in the flow cell and incubated for 1 min. 2 mm ATP was introduced in the chamber to release the actin filaments and to make the active motor heads accessible. ATP was washed out with assay buffer, tetramethylrhodamine-labeled F-actin was incubated for 1 min and washed to remove excess filaments, and finally the chamber was infused with buffer containing 2 mm MgATP and antibleach system (18 μg/ml catalase, 0.1 mg/ml glucose oxidase, 10 mg/ml d-glucose, and 10 mm DTT) to initiate F-actin motility. Images were acquired with a time resolution of 200 ms (i.e. 5 frames/s) using a custom-made objective-type total internal reflection fluorescence microscope. Actin filament gliding speed was analyzed with the Manual Tracking plugin from ImageJ.
Three-bead assay with optical tweezers
For the assay, flow cells with ∼15-μl chamber volumes were assembled using a coverslip with nitrocellulose-coated beads. Glass microspheres (1–1.5 μm) suspended in 0.1% nitrocellulose in amyl acetate were applied to 18 × 18-mm coverslips. All of the dilutions of biotin-actin filaments and myosin S1 were made in reaction buffer containing 25 mm KCl, 25 mm Hepes (pH 7.6), 4 mm MgCl2, and 1% β-mercaptoethanol. For the experiment, the chamber was prepared as follows. 1) Flow cells were first incubated with 1 μg/ml myosin S1 for 1 min. 2) This was followed by a wash with 1 mg/ml BSA and further incubation for 1 min to block the surface. 3) Finally, the reaction mixture, containing 0.8–1-μm neutravidin-coated polystyrene beads and 1–2 nm biotinylated actin filaments (60), was flowed in with 10 μm ATP, ATP-regenerating system (0.01 mm creatine phosphate and 0.01 unit creating kinase), and deoxygenating system (1 mg/ml catalase, 1 mg/ml glucose oxidase, 2 mg/ml glucose, and 1% β-mercaptoethanol). As shown in the schematic view in Fig. 4A, prestretched, biotinylated actin filament was held between the two optically trapped beads via a neutravidin-biotin link forming a dumbbell, such that the actin filament formed a connection between two beads. The approximate distance between the two trapped beads was 10 μm. Low-compliance links between the trapped beads and the filament were adjusted such that the ratio of the position variance during free and bound periods was 5–10, as described by Smith et al. (49). All of the experiments were performed at room temperature of ∼22 °C. WT and chimeric motor S1 were immobilized directly on a nitrocellulose-coated surface to have comparable conditions for the single molecules. The actin dumbbell was brought in contact with the myosin bead, and the AM interaction events were registered as a reduction in free Brownian noise of the two beads. The two bead positions were precisely detected with a quadrant photodetector, recorded, and analyzed.
Data traces were collected at a sampling rate of 10,000 Hz and filtered at 5000 Hz. To improve the time resolution and detect short-lived AM binding events, a high-frequency triangular wave of ∼600 Hz was applied to one of the traps as described by Veigel et al. (61). Matlab routines were employed to evaluate data records for AM interaction lifetime, ton, and stroke size of motors.
Single-myosin molecule interaction with actin filaments in optical trapping measurements
To ensure that each data record was derived from an intermittent interaction between single myosin head and actin, myosin density on the bead surface was adjusted by diluting the myosin solution. In our measurements, typically, one bead was found to interact with dumbbell among 8–10 beads scanned for the presence of motor on the bead. This measure minimized the likelihood of multiple molecules simultaneously interacting with the actin filaments. From the binomial probability distribution, in our measurements, chances of the presence of two motors per bead are <1%, and chances of one molecule per bead are ∼9%. From a total of 126 beads we analyzed for AM-binding events, statistically, 0.4 beads may have two motors. This estimation, however, does not take into account the motors that are inaccessible to actin filament due to the positional limitation on the bead. Therefore, the probability of multiple motors interacting simultaneously with an actin filament is even lower.
Statistical analysis
Binomial probability distribution analysis was used to estimate the chances of more than one molecule interacting with actin filaments in optical trap measurements. Unpaired t test was used to calculate the statistical differences in the gliding velocities, single-motor stroke size, and stiffness of the native versus chimeric motors. The nonparametric Mann–Whitney U test was used to calculate the statistical differences in the duration of AM interaction events, ton, for the WT and chimeric motors. The statistical tests used to calculate statistical significance are included throughout in relevant sections.
All of the experimental protocols were performed in accordance with guidelines for good scientific practices and approved by Hannover Medical School.
Data availability
All data are available within the article.
Author contributions
A. N. and M. A. data curation; A. N., T. W., P. F., W. S., I. C., G. T., and M. A. formal analysis; A. N. and M. A. investigation; A. N., T. W., P. F., W. S., G. T., and M. A. writing-review and editing; T. W., P. F., W. S., I. C., and G. T. methodology; W. S., I. C., and G. T. software; M. A. conceptualization; M. A. supervision; M. A. funding acquisition; M. A. writing-original draft; M. A. project administration.
Supplementary Material
Acknowledgments
We thank the late Prof. Bernhard Brenner. We thank Dr. Atsuko Iwane for a kind gift of BDTC-chicken gizzard RLC construct to Dr. Walter Steffen. We thank Petra Uta and Stefanie Nedel for the technical assistance in protein production and purification, Prof. Theresia Kraft and Bogdan Iorga for critical comments on the manuscript, and Dr. Ante Radocaj for comments on statistical analysis.
This research was supported by Deutsche Forschungsgemeinschaft (DFG) Grant AM/507/1-1 (to M. A.). The authors declare that they have no conflicts of interest with the contents of this article.
We dedicate this work to the late Prof. Bernhard Brenner.
This article contains supporting Figs. S1 and S2 and Movies S1 and S2.
- MHC
- myosin heavy chain
- RLC
- regulatory light chain
- LC
- light chain
- MLC
- myosin light chain
- ELC
- essential light chain
- MLC1f
- myosin light chain 1 fast
- MLC2f
- myosin light chain 2 fast
- MLC2v
- myosin light chain 2 ventricular
- MLC2s
- myosin light chain 2 slow
- S1 and S2
- subfragment 1 and 2, respectively
- SMM
- smooth muscle myosin
- cgmRLC
- chicken gizzard smooth muscle myosin RLC
- AM
- actomyosin
- Vmax
- maximum velocity
- MLC2B
- myosin light chain 2 fast
- pN
- piconewtons
- aa
- amino acids
- BDTC
- biotin-dependent transcarboxylase
- mantADP
- 2′,3′-O-(N′-methylanthraniloyl)-ADP.
References
- 1. Lowey S., and Risby D. (1971) Light chains from fast and slow muscle myosins. Nature 234, 81–85 10.1038/234081a0 [DOI] [PubMed] [Google Scholar]
- 2. Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., and Holden H. M. (1993) Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261, 50–58 10.1126/science.8316857 [DOI] [PubMed] [Google Scholar]
- 3. Uyeda T. Q., Abramson P. D., and Spudich J. A. (1996) The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. U.S.A. 93, 4459–4464 10.1073/pnas.93.9.4459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lowey S., and Trybus K. M. (1995) Role of skeletal and smooth muscle myosin light chains. Biophys J 68, 120S–126S; discussion 126S–127S [PMC free article] [PubMed] [Google Scholar]
- 5. Schiaffino S., and Reggiani C. (1994) Myosin isoforms in mammalian skeletal muscle. J. Appl. Physiol. 77, 493–501 10.1152/jappl.1994.77.2.493 [DOI] [PubMed] [Google Scholar]
- 6. Iorga B., Adamek N., and Geeves M. A. (2007) The slow skeletal muscle isoform of myosin shows kinetic features common to smooth and non-muscle myosins. J. Biol. Chem. 282, 3559–3570 10.1074/jbc.M608191200 [DOI] [PubMed] [Google Scholar]
- 7. Sheikh F., Lyon R. C., and Chen J. (2015) Functions of myosin light chain-2 (MYL2) in cardiac muscle and disease. Gene 569, 14–20 10.1016/j.gene.2015.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kampourakis T., and Irving M. (2015) Phosphorylation of myosin regulatory light chain controls myosin head conformation in cardiac muscle. J. Mol. Cell Cardiol. 85, 199–206 10.1016/j.yjmcc.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kampourakis T., Sun Y. B., and Irving M. (2016) Myosin light chain phosphorylation enhances contraction of heart muscle via structural changes in both thick and thin filaments. Proc. Natl. Acad. Sci. U.S.A. 113, E3039–E3047 10.1073/pnas.1602776113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scruggs S. B., and Solaro R. J. (2011) The significance of regulatory light chain phosphorylation in cardiac physiology. Arch. Biochem. Biophys. 510, 129–134 10.1016/j.abb.2011.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morano I. (1992) Effects of different expression and posttranslational modifications of myosin light chains on contractility of skinned human cardiac fibers. Basic Res. Cardiol. 87, 129–141 10.1007/978-3-642-72474-9_11 [DOI] [PubMed] [Google Scholar]
- 12. Persechini A., Stull J. T., and Cooke R. (1985) The effect of myosin phosphorylation on the contractile properties of skinned rabbit skeletal muscle fibers. J. Biol. Chem. 260, 7951–7954 [PubMed] [Google Scholar]
- 13. Sweeney H. L., and Stull J. T. (1986) Phosphorylation of myosin in permeabilized mammalian cardiac and skeletal muscle cells. Am. J. Physiol. 250, C657–C660 10.1152/ajpcell.1986.250.4.C657 [DOI] [PubMed] [Google Scholar]
- 14. Andruchov O., Andruchova O., Wang Y., and Galler S. (2006) Dependence of cross-bridge kinetics on myosin light chain isoforms in rabbit and rat skeletal muscle fibres. J Physiol. 571, 231–242 10.1113/jphysiol.2005.099770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reiser P. J., Moss R. L., Giulian G. G., and Greaser M. L. (1985) Shortening velocity in single fibers from adult rabbit soleus muscles is correlated with myosin heavy chain composition. J. Biol. Chem. 260, 9077–9080 [PubMed] [Google Scholar]
- 16. Larsson L., and Moss R. L. (1993) Maximum velocity of shortening in relation to myosin isoform composition in single fibres from human skeletal muscles. J. Physiol. 472, 595–614 10.1113/jphysiol.1993.sp019964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salviati G., Betto R., and Danieli Betto D. (1982) Polymorphism of myofibrillar proteins of rabbit skeletal-muscle fibres. An electrophoretic study of single fibres. Biochem. J. 207, 261–272 10.1042/bj2070261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Staron R. S., and Pette D. (1987) Nonuniform myosin expression along single fibers of chronically stimulated and contralateral rabbit tibialis anterior muscles. Pflugers Arch. 409, 67–73 10.1007/BF00584751 [DOI] [PubMed] [Google Scholar]
- 19. Stephenson G. M. (2001) Hybrid skeletal muscle fibres: a rare or common phenomenon? Clin. Exp. Pharmacol. Physiol. 28, 692–702 10.1046/j.1440-1681.2001.03505.x [DOI] [PubMed] [Google Scholar]
- 20. Iwane A. H., Kitamura K., Tokunaga M., and Yanagida T. (1997) Myosin subfragment-1 is fully equipped with factors essential for motor function. Biochem. Biophys. Res. Commun. 230, 76–80 10.1006/bbrc.1996.5861 [DOI] [PubMed] [Google Scholar]
- 21. Warshaw D. M., Desrosiers J. M., Work S. S., and Trybus K. M. (1990) Smooth muscle myosin cross-bridge interactions modulate actin filament sliding velocity in vitro. J. Cell Biol. 111, 453–463 10.1083/jcb.111.2.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harris D. E., and Warshaw D. M. (1993) Smooth and skeletal muscle actin are mechanically indistinguishable in the in vitro motility assay. Circ. Res. 72, 219–224 10.1161/01.RES.72.1.219 [DOI] [PubMed] [Google Scholar]
- 23. Cuda G., Pate E., Cooke R., and Sellers J. R. (1997) In vitro actin filament sliding velocities produced by mixtures of different types of myosin. Biophys. J. 72, 1767–1779 10.1016/S0006-3495(97)78823-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Veigel C., Molloy J. E., Schmitz S., and Kendrick-Jones J. (2003) Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat. Cell Biol. 5, 980–986 10.1038/ncb1060 [DOI] [PubMed] [Google Scholar]
- 25. Siemankowski R. F., and White H. D. (1984) Kinetics of the interaction between actin, ADP, and cardiac myosin-S1. J. Biol. Chem. 259, 5045–5053 [PubMed] [Google Scholar]
- 26. Weiss S., Rossi R., Pellegrino M. A., Bottinelli R., and Geeves M. A. (2001) Differing ADP release rates from myosin heavy chain isoforms define the shortening velocity of skeletal muscle fibers. J. Biol. Chem. 276, 45902–45908 10.1074/jbc.M107434200 [DOI] [PubMed] [Google Scholar]
- 27. Harris D. E., and Warshaw D. M. (1993) Smooth and skeletal muscle myosin both exhibit low duty cycles at zero load in vitro. J. Biol. Chem. 268, 14764–14768 [PubMed] [Google Scholar]
- 28. Uyeda T. Q., Kron S. J., and Spudich J. A. (1990) Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 214, 699–710 10.1016/0022-2836(90)90287-V [DOI] [PubMed] [Google Scholar]
- 29. Stein L. A., Greene L. E., Chock P. B., and Eisenberg E. (1985) Rate-limiting step in the actomyosin adenosinetriphosphatase cycle: studies with myosin subfragment 1 cross-linked to actin. Biochemistry 24, 1357–1363 10.1021/bi00327a013 [DOI] [PubMed] [Google Scholar]
- 30. Lionne C., Iorga B., Candau R., Piroddi N., Webb M. R., Belus A., Travers F., and Barman T. (2002) Evidence that phosphate release is the rate-limiting step on the overall ATPase of psoas myofibrils prevented from shortening by chemical cross-linking. Biochemistry 41, 13297–13308 10.1021/bi0260278 [DOI] [PubMed] [Google Scholar]
- 31. Amrute-Nayak M., Nayak A., Steffen W., Tsiavaliaris G., Scholz T., and Brenner B. (2019) Transformation of the nonprocessive fast skeletal myosin II into a processive motor. Small 15, e1804313 10.1002/smll.201804313 [DOI] [PubMed] [Google Scholar]
- 32. Ruppel K. M., and Spudich J. A. (1996) Structure-function studies of the myosin motor domain: importance of the 50-kDa cleft. Mol. Biol. Cell 7, 1123–1136 10.1091/mbc.7.7.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsiavaliaris G., Fujita-Becker S., Batra R., Levitsky D. I., Kull F. J., Geeves M. A., and Manstein D. J. (2002) Mutations in the relay loop region result in dominant-negative inhibition of myosin II function in Dictyostelium. EMBO Rep. 3, 1099–1105 10.1093/embo-reports/kvf214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liao W., Elfrink K., and Bähler M. (2010) Head of myosin IX binds calmodulin and moves processively toward the plus-end of actin filaments. J. Biol. Chem. 285, 24933–24942 10.1074/jbc.M110.101105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haithcock J., Billington N., Choi K., Fordham J., Sellers J. R., Stafford W. F., White H., and Forgacs E. (2011) The kinetic mechanism of mouse myosin VIIA. J. Biol. Chem. 286, 8819–8828 10.1074/jbc.M110.163592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Steffen W., Smith D., Simmons R., and Sleep J. (2001) Mapping the actin filament with myosin. Proc. Natl. Acad. Sci. U.S.A. 98, 14949–14954 10.1073/pnas.261560698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muretta J. M., Rohde J. A., Johnsrud D. O., Cornea S., and Thomas D. D. (2015) Direct real-time detection of the structural and biochemical events in the myosin power stroke. Proc. Natl. Acad. Sci. U.S.A. 112, 14272–14277 10.1073/pnas.1514859112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Woody M. S., Winkelmann D. A., Capitanio M., Ostap E. M., and Goldman Y. E. (2019) Single molecule mechanics resolves the earliest events in force generation by cardiac myosin. Elife 8, e49266 10.7554/eLife.49266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Capitanio M., Canepari M., Cacciafesta P., Lombardi V., Cicchi R., Maffei M., Pavone F. S., and Bottinelli R. (2006) Two independent mechanical events in the interaction cycle of skeletal muscle myosin with actin. Proc. Natl. Acad. Sci. U.S.A. 103, 87–92 10.1073/pnas.0506830102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Batters C., and Veigel C. (2011) Using optical tweezers to study the fine details of myosin ATPase mechanochemical cycle. Methods Mol. Biol. 778, 97–109 10.1007/978-1-61779-261-8_7 [DOI] [PubMed] [Google Scholar]
- 41. Karatzaferi C., Adamek N., and Geeves M. A. (2017) Modulators of actin-myosin dissociation: basis for muscle type functional differences during fatigue. Am. J. Physiol. Cell Physiol. 313, C644–C654 10.1152/ajpcell.00023.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ruff C., Furch M., Brenner B., Manstein D. J., and Meyhöfer E. (2001) Single-molecule tracking of myosins with genetically engineered amplifier domains. Nat. Struct. Biol. 8, 226–229 10.1038/84962 [DOI] [PubMed] [Google Scholar]
- 43. Molloy J. E., Burns J. E., Kendrick-Jones J., Tregear R. T., and White D. C. (1995) Movement and force produced by a single myosin head. Nature 378, 209–212 10.1038/378209a0 [DOI] [PubMed] [Google Scholar]
- 44. Huxley A. F., and Simmons R. M. (1971) Proposed mechanism of force generation in striated muscle. Nature 233, 533–538 10.1038/233533a0 [DOI] [PubMed] [Google Scholar]
- 45. Reconditi M., Linari M., Lucii L., Stewart A., Sun Y. B., Boesecke P., Narayanan T., Fischetti R. F., Irving T., Piazzesi G., Irving M., and Lombardi V. (2004) The myosin motor in muscle generates a smaller and slower working stroke at higher load. Nature 428, 578–581 10.1038/nature02380 [DOI] [PubMed] [Google Scholar]
- 46. Veigel C., Schmitz S., Wang F., and Sellers J. R. (2005) Load-dependent kinetics of myosin-V can explain its high processivity. Nat. Cell Biol. 7, 861–869 10.1038/ncb1287 [DOI] [PubMed] [Google Scholar]
- 47. Steffen W., Lewalle A., and Sleep J. (2006) Optical tweezers: Application to the study of motor proteins. Cell Biology, Vol. 3, pp. 37–45, Elsevier Acad. Press, USA [Google Scholar]
- 48. Lewalle A., Steffen W., Stevenson O., Ouyang Z., and Sleep J. (2008) Single-molecule measurement of the stiffness of the rigor myosin head. Biophys. J. 94, 2160–2169 10.1529/biophysj.107.119396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith D. A., Steffen W., Simmons R. M., and Sleep J. (2001) Hidden-Markov methods for the analysis of single-molecule actomyosin displacement data: the variance-Hidden-Markov method. Biophys. J. 81, 2795–2816 10.1016/S0006-3495(01)75922-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Percario V., Boncompagni S., Protasi F., Pertici I., Pinzauti F., and Caremani M. (2018) Mechanical parameters of the molecular motor myosin II determined in permeabilised fibres from slow and fast skeletal muscles of the rabbit. J. Physiol. 596, 1243–1257 10.1113/JP275404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Szczesna D., Ghosh D., Li Q., Gomes A. V., Guzman G., Arana C., Zhi G., Stull J. T., and Potter J. D. (2001) Familial hypertrophic cardiomyopathy mutations in the regulatory light chains of myosin affect their structure, Ca2+ binding, and phosphorylation. J. Biol. Chem. 276, 7086–7092 10.1074/jbc.M009823200 [DOI] [PubMed] [Google Scholar]
- 52. Iorga B., Schwanke K., Weber N., Wendland M., Greten S., Piep B., Dos Remedios C. G., Martin U., Zweigerdt R., Kraft T., and Brenner B. (2017) Differences in contractile function of myofibrils within human embryonic stem cell-derived cardiomyocytes vs. adult ventricular myofibrils are related to distinct sarcomeric protein isoforms. Front. Physiol. 8, 1111 10.3389/fpls.2017.01111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Amrute-Nayak M., Antognozzi M., Scholz T., Kojima H., and Brenner B. (2008) Inorganic phosphate binds to the empty nucleotide binding pocket of conventional myosin II. J. Biol. Chem. 283, 3773–3781 10.1074/jbc.M706779200 [DOI] [PubMed] [Google Scholar]
- 54. Thedinga E., Karim N., Kraft T., and Brenner B. (1999) A single-fiber in vitro motility assay: in vitro sliding velocity of F-actin vs. unloaded shortening velocity in skinned muscle fibers. J. Muscle Res. Cell Motil. 20, 785–796 10.1023/A:1005658825375 [DOI] [PubMed] [Google Scholar]
- 55. Margossian S. S., and Lowey S. (1982) Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 85, 55–71 10.1016/0076-6879(82)85009-X [DOI] [PubMed] [Google Scholar]
- 56. Trybus K. M., and Chatman T. A. (1993) Chimeric regulatory light chains as probes of smooth muscle myosin function. J. Biol. Chem. 268, 4412–4419 [PubMed] [Google Scholar]
- 57. Rajasekharan K. N., Morita J. I., Mayadevi M., Ikebe M., and Burke M. (1991) Formation and properties of smooth muscle myosin 20-kDa light chain-skeletal muscle myosin hybrids and photocrosslinking from the maleimidylbenzophenone-labeled light chain to the heavy chain. Arch. Biochem. Biophys. 288, 584–590 10.1016/0003-9861(91)90240-J [DOI] [PubMed] [Google Scholar]
- 58. Katoh T., and Lowey S. (1989) Mapping myosin light chains by immunoelectron microscopy. Use of anti-fluorescyl antibodies as structural probes. J. Cell Biol. 109, 1549–1560 10.1083/jcb.109.4.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brune M., Hunter J. L., Corrie J. E., and Webb M. R. (1994) Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33, 8262–8271 10.1021/bi00193a013 [DOI] [PubMed] [Google Scholar]
- 60. Ishijima A., Kojima H., Funatsu T., Tokunaga M., Higuchi H., Tanaka H., and Yanagida T. (1998) Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell 92, 161–171 10.1016/S0092-8674(00)80911-3 [DOI] [PubMed] [Google Scholar]
- 61. Veigel C., Wang F., Bartoo M. L., Sellers J. R., and Molloy J. E. (2002) The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 4, 59–65 10.1038/ncb732 [DOI] [PubMed] [Google Scholar]
- 62. Risal D., Gourinath S., Himmel D. M., Szent-Györgyi A. G., and Cohen C. (2004) Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding. Proc. Natl. Acad. Sci. U.S.A. 101, 8930–8935 10.1073/pnas.0403002101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available within the article.