Figure 1.
Analysis of Protein Expression Following Genome Editing
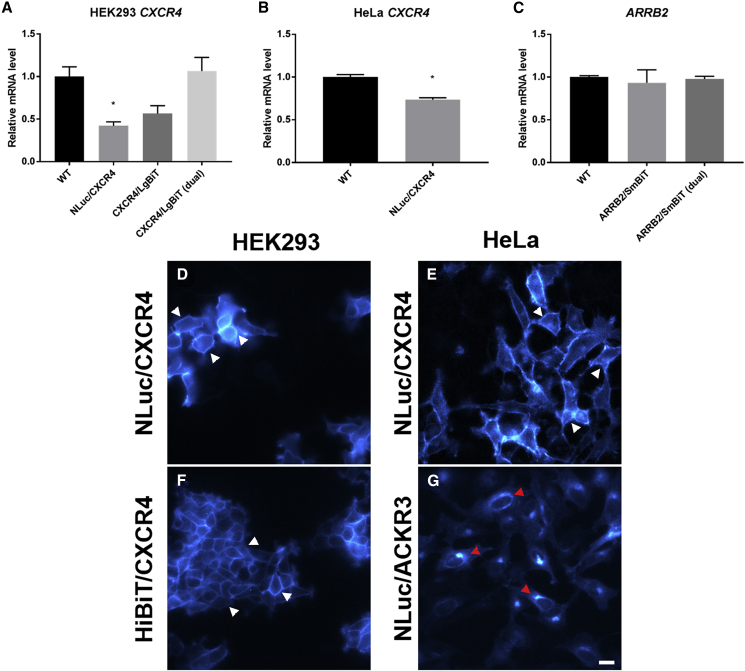
(A) CXCR4 mRNA expression in wild-type HEK293 cells or HEK293 clones expressing genome-edited NLuc/CXCR4, CXCR4/LgBiT, or CXCR4/LgBiT and ARRB2/SmBiT (dual).
(B) CXCR4 mRNA expression in wild-type HeLa cells or HeLa clones expressing genome-edited NLuc/CXCR4.
(C) ARRB2 mRNA expression in wild-type HEK293 cells or HEK293 clones expressing genome-edited ARRB2/SmBiT, or ARRB2/SmBiT and CXCR4/LgBiT (dual). Relative mRNA level, normalized to BM2 expression. Bars represent mean ± SEM of three cell passages of a single clone performed in triplicate.
(D–G) Visualization of genome-edited receptor localization in HEK293 and HeLa cells using a bioluminescence LV200 Olympus microscope. (D) HEK293 and (E) HeLa cells expressing genome-edited NLuc/CXCR4, (F) HEK293 cells expressing genome-edited HiBiT/CXCR4 complemented with LgBiT and (G) HeLa cells expressing genome-edited NLuc/ACKR3. White arrow heads (D–F) indicate predominant expression at the plasma membrane of luciferase-tagged CXCR4, red arrow heads (G) indicate NLuc/ACKR3 expression in cytosolic compartments. Images were acquired by capturing total luminescence for 90 s.
Scale bar represents 20 μm. See Figure S1.