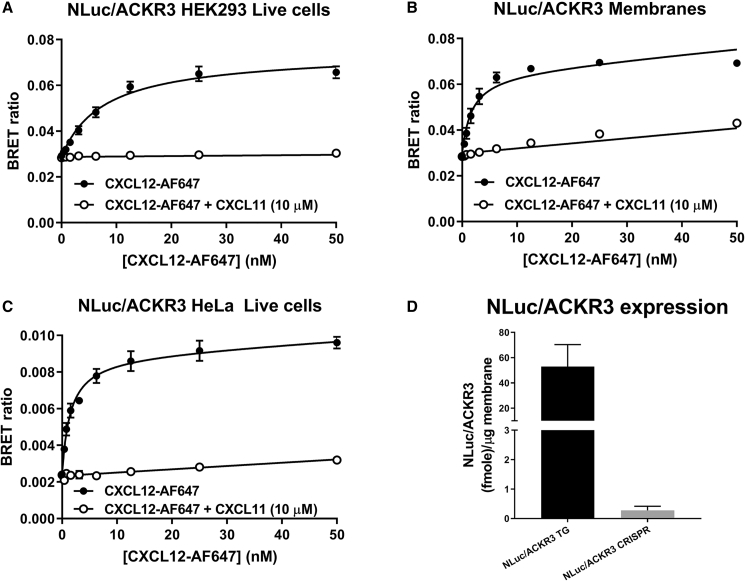
Figure 3.
Determination of the Binding Affinity of CXCL12-AF647 at NLuc/ACKR3
(A–C) NanoBRET saturation ligand binding curves obtained in (A) live HEK293 cells exogenously expressing NLuc/ACKR3, (B) membrane preparations from HEK293 cells exogenously expressing NLuc/ACKR3, and (C) live HeLa cells expressing genome-edited NLuc/ACKR3. Cells or membranes were incubated with increasing concentrations of CXCL12-AF647 in the absence (black circles) or presence (white circles) of CXCL11 (10 μM) for 1 h at 37°C. Data shown are mean ± SEM and are representative of five (A), four (B), and six (C) experiments performed in duplicate.
(D) Quantification of NLuc/ACKR3 expression by linear regression, as described in the STAR Methods, using membrane preparations made from HEK293 cells exogenously expressing NLuc/ACKR3 (NLuc/ACKR3 TG, black bar) or HeLa cells expressing genome-edited NLuc/ACKR3 (NLuc/ACKR3 CRISPR, gray bar). Data shown are mean ± SEM of five individual experiments performed in triplicate.