Figure 1.
Elg1 Removal Compensates for Absence of Ctf18
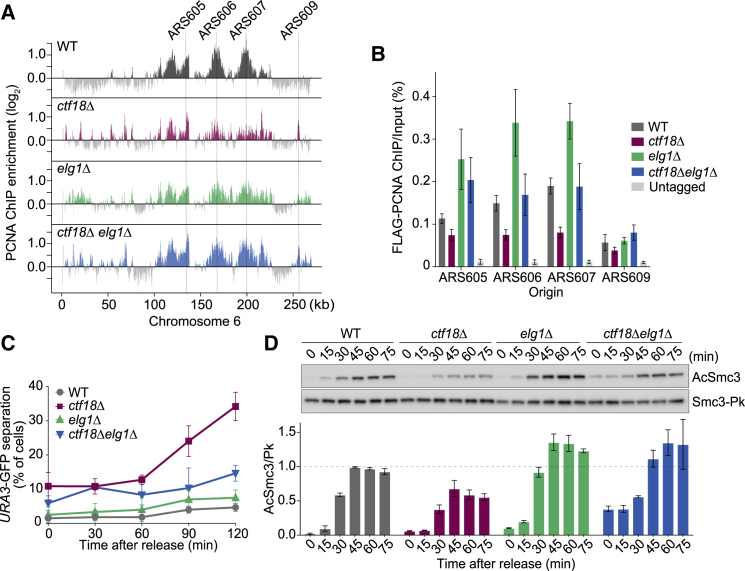
(A) PCNA distributions in the absence of Ctf18 and/or Elg1. Cells were synchronized in G1 and released into HU-containing medium. PCNA chromatin immunoprecipitates were hybridized to Affymetrix GeneChip S. cerevisiae Tiling 1.0R arrays. Signal intensities, relative to a whole-genome DNA sample, are shown along chromosome 6. Replication origins chosen for subsequent quantitative analyses are indicated.
(B) As in (A), but chromatin immunoprecipitates from N-terminally FLAG epitope-tagged PCNA were analyzed using quantitative real-time PCR using primer pairs at an early (ARS605, 606, and 607) and a late firing (ARS609) replication origin. Means ± SE from three independent experiments are shown.
(C) Cells of the indicated genotypes were synchronized in G1 and released into nocodazole-containing medium to induce a mitotic arrest. Sister chromatid cohesion was assessed at the GFP-marked URA3 locus at indicated time points. Means ± SE from three independent experiments are shown.
(D) As in (C), but Smc3 acetylation was monitored by western blotting using an acetyl-Smc3-specific (AcSmc3) antibody. Total Smc3 levels were detected by its Pk epitope and served as a loading control. The AcSmc3/Smc3-Pk ratio was normalized to that in wild-type cells at 45 min. Means ± SE from three independent experiments are shown.
See Figures S1A and S1B for confirmation of Ctf18 binding and PCNA loading at forks progressing through undisturbed S phase and Figures S2A–S2E for experiments separating Ctf18’s function in sister chromatid cohesion and the replication checkpoint.