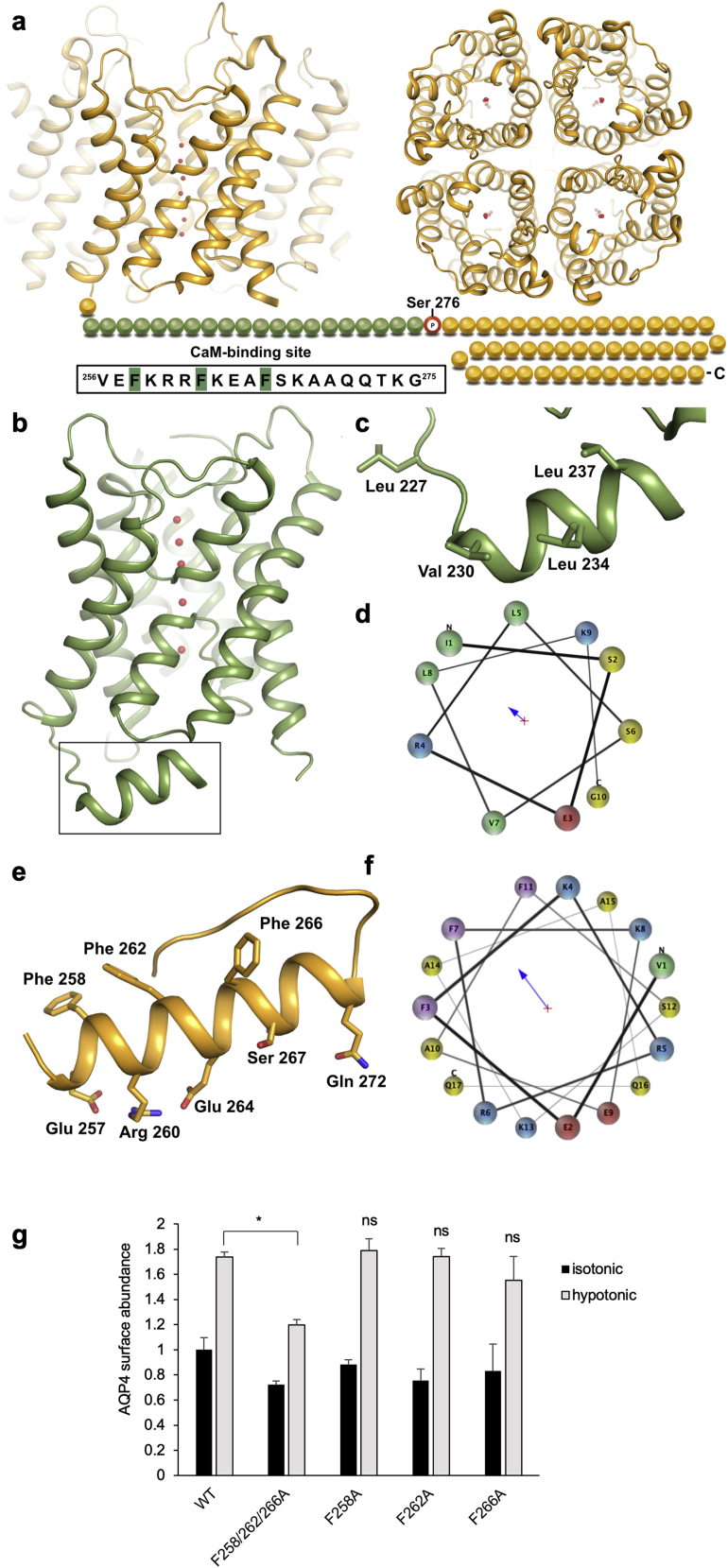
Figure S4.
The Predicted AQP4 CBD and Its Comparison with the CBD in the AQP0 Crystal Structure
a, Crystal structure of the human AQP4 tetramer viewed from the side of the membrane and from the extracellular side. The carboxyl terminus, for which there is no structural information, is shown as beads. The sequence of the predicted CaM-binding domain (green beads) is shown in the box with hydrophobic residues highlighted in green. The phosphorylation site at Ser 276 is highlighted with a red circle; b, Crystal structure of bovine AQP0 (PDB code 1YMG) showing the carboxy-terminal helix (black box) which harbors the CaM-binding domain of AQP0; c, Zoom-in on the CaM binding domain with residues involved in binding shown in stick representation; d, Helix wheel representation of the AQP0 carboxy-terminal helix showing its amphipathic character. Colors indicate residue types as follows: hydrophobic-green, basic-blue, acidic-red and polar-yellow; e, Top scoring structural model of the predicted AQP4 CaM-binding site generated by PEP-FOLD3. Hydrophobic and charged/polar residues on either side of the predicted helix are shown in stick representation; f, Helical wheel representation of the predicted helix in panel e showing its amphipathic character. Colors indicate residue types as follows: aromatic-purple, hydrophobic-green, basic-blue, acidic-red and polar-yellow; g, Hypotonicity-induced translocation of AQP4 in HEK293 cells is abrogated by a triple F258/262/266A mutation, but not by the corresponding single point mutations. ∗p < 0.05 by ANOVA followed by Bonferroni-corrected t test, ns denotes p > 0.05 (see Table S2 for p values). Related to Figure 3.