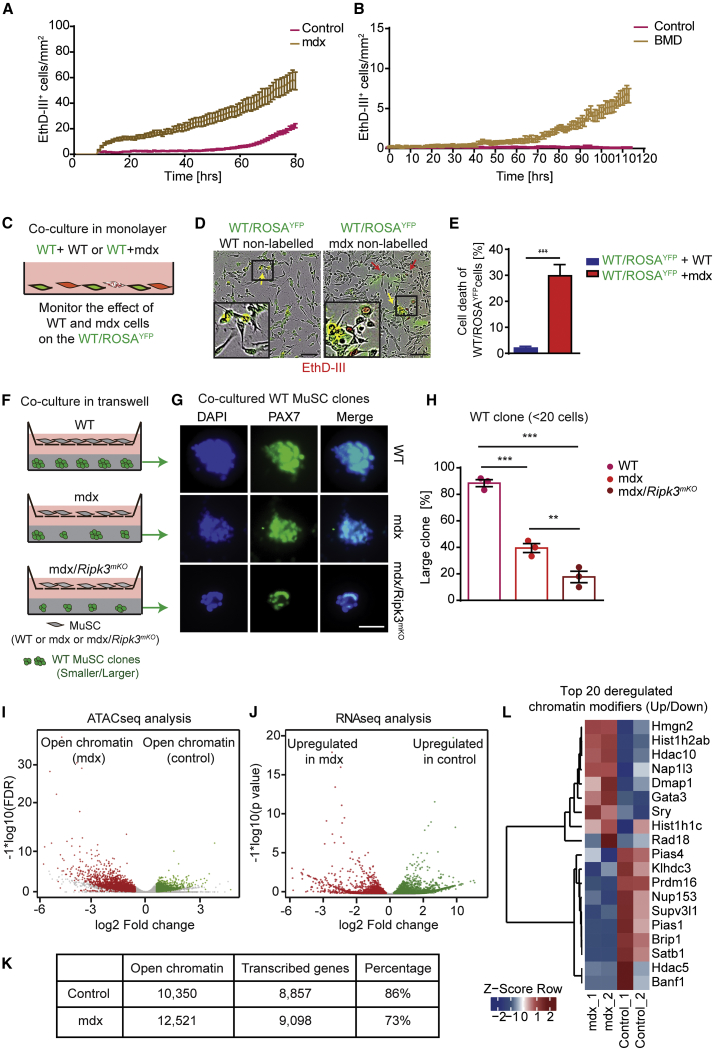
Figure 3.
Predisposition of MuSCs from Dystrophic Muscles for Necroptosis Reduces Adverse Crosstalk among mdx Muscle Stem Cells and Is Associated with Chromatin Reorganization
(A) EthD-III incorporation rates of MuSCs isolated from control and mdx mice in vitro.
(B) EthD-III incorporation rates of MuSCs isolated from healthy individuals and BMD patients in vitro (n = 3 for each group in both A and B).
(C) Schematic of co-culture setup with YFP+ WT MuSCs in direct monolayer co-culture with either YFP− WT or YFP−mdx MuSCs.
(D) Representative images of co-cultured MuSCs (insets are magnifications of indicated boxes). Scale bar, 100 μm.
(E) Quantification of YFP+ WT MuSCs incorporating EthD-III at endpoints as in (D) (n = 3 for each group).
(F) Schematic of transwell-assay setup. WT, mdx, and mdx/Ripk3mKO MuSCs are grown in the upper transwell; in the lower well, single WT MuSCs are grown in Matrigel at clonal density.
(G and H) Representative images of MuSC colony sizes after 4-day culture in Matrigel (G), quantified in (H). n = 3 for each group. Scale bar in (G): 100 μm.
(I) Volcano plot of ATAC-seq data showing increase of chromatin accessibility in mdx MuSCs (red) compared with control MuSCs (green). n = 2 or 3 for each group.
(J) Volcano plot of RNA-seq data showing increase of transcriptional upregulation in mdx MuSCs (red) compared with control MuSCs (green). n = 2 or 3 for each group.
(K) Correlation scores between chromatin accessibility from (I) and actively transcribed genes from (J).
(L) Heatmap representation of genes coding for chromatin remodelers with more (red) and less (blue) accessible chromatin in MuSCs derived from control and mdx mice (n = 2 for each group).
Statistical analysis: ∗∗p < 0.01 and ∗∗∗p < 0.005, two-way ANOVA followed by Bonferroni post-test with alpha = 5%. All analyses indicated across the experiments were biological replicates unless otherwise stated.