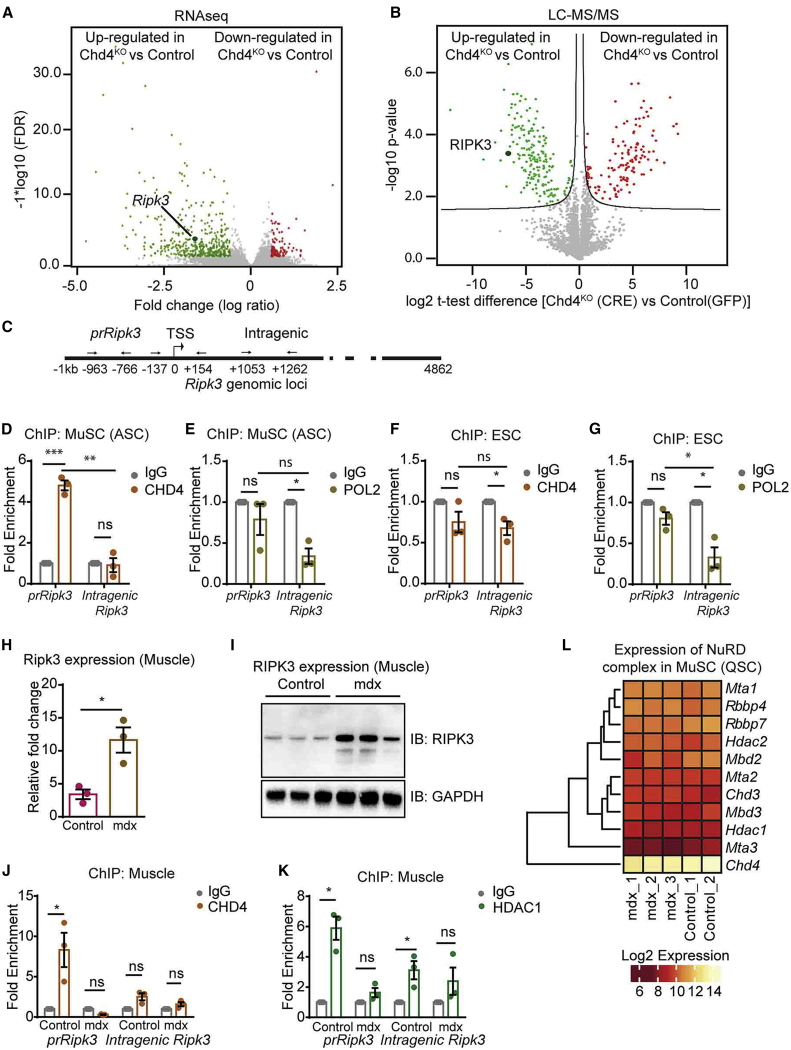
Figure 5.
CHD4 Directly Suppresses Activation of Ripk3
(A and B) Volcano plots of RNA-seq (A) and proteomics data (B) visualizing significantly downregulated (red points) or upregulated (green points) transcripts and proteins after adeno-Cre-mediated inactivation of Chd4 in MuSCs from Chd4loxP/loxP mice (n = 4).
(C) Schematic representation of the locations of primers used for ChIP-qPCR to detect binding of Chd4 to the Ripk3 gene.
(D and E) qRT-PCR ChIP analyses of CHD4 binding (D) and POL II binding (E) in activated MuSCs (ASC).
(F and G) qRT-PCR ChIP analyses of CHD4 binding (F) and POL II binding (G) in embryonic stem cells (ESCs). IgG served as a negative control (n = 3 independent technical replicates for each group).
(H and I) qRT-PCR (H) and western blot (I) analyses of Ripk3 expression in control and mdx muscle tissues (n = 3).
(J and K) ChIP-qPCR analysis of CHD4 binding (J) and HDAC1 binding (K) to the promoter and intragenic regions of the Ripk3 gene in skeletal muscles from control and mdx mice (n = 3 independent technical replicates for each group).
(L) Heatmap representing gene expression of members of the NuRD complex (log2 expression) in quiescent MuSCs (QSC) from control (n = 2) and mdx (n = 3) mice on the basis of RNA-seq data.
Statiscal analysis: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005, two-way ANOVA followed by Bonferroni post-test with alpha = 5%). All analyses indicated across the experiments were biological replicates unless otherwise stated.