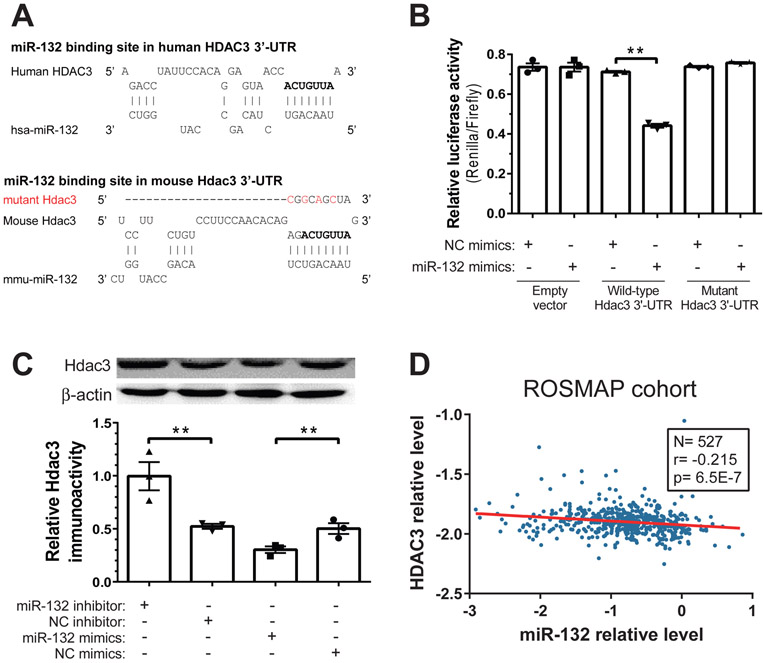
Figure 5. HDAC3 is a direct target of miR-132-3p.
(A) The predicted binding site of miR-132-3p in the 3’-UTR of HDAC3 mRNA in human and mouse, which is also observed by CLIP-Seq datasets. The conserved seed binding site is shown in bold. As a negative control, this seed binding region was disrupted by four mutations (labeled in red) in the mutant vector. (B) The effects of miR-132-3p on HDAC3 3’-UTR were examined by co-transfection of the dual-luciferase reporter system with either miR-132-3p mimics or negative control (NC) mimics. Overexpression of miR-132-3p specifically downregulated the Renilla reporter for HDAC3 3’-UTR but not for the mutant version. N=3. (C) The effect of miR-132-3p on HDAC3 protein level in neurons. Primary mouse cortical neurons were transfected with miR-132-3p inhibitor, NC inhibitor, mimics or NC mimics, and Aβ1-42 (1 μM) was added 24 hr later. Total protein was extracted after another 24 hr, and the HDAC3 protein was quantified by Western blot, with β-actin as the reference protein. N=3. (D) HDAC3 mRNA relative levels are negatively correlated with miR-132 relative levels in postmortem brain tissues from the ROSMAP cohort (N=527). miR-132 microarray data were normalized to miR-99a, and HDAC3 RNAseq data were normalized to GAPDH following log-transformation. Spearman correlation, p<0.001. All data are means ± S.E.M; *, p<0.05; **, p<0.01