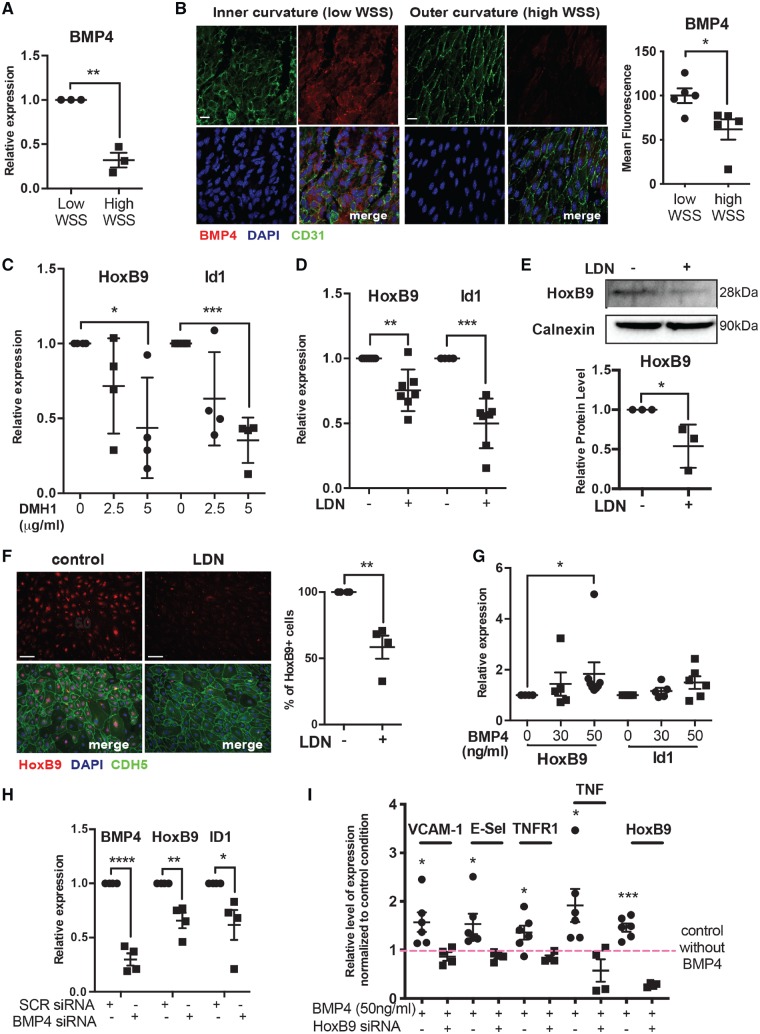
Figure 5.
BMP4 induces HoxB9 in response to low shear stress. (A) HUVEC were cultured in 6-well plates and exposed to orbital shaking to generate WSS. After 72 h, cells were isolated from the centre (low WSS) or the periphery (high WSS) of the well. BMP4 mRNA levels were quantified by qRT-PCR using gene-specific primers. N = 3 experiments. (B) The expression of BMP4 was quantified at the inner curvature (low WSS) and outer curvature (high WSS) of the murine aortic arch by en face staining (red). EC were identified using anti-CD31 antibodies (green) and nuclei were co-stained using TOPRO3 (blue). N = 3. (C–F) HUVEC were treated with DMH1 or LDN and then exposed to low WSS for 72 h. Bar = 10 μm. (C and D) mRNA levels of HoxB9 or ID1 were quantified by qRT-PCR using gene-specific primers. N = 4–7 experiments. (E) The expression levels of HoxB9 were assessed by Western blotting using specific antibodies and anti-Calnexin was used to control for total protein levels. N = 4 experiments. (F) HoxB9 expression was assessed by immunofluorescent staining (red). EC were identified using anti-CDH5 antibodies (green) and nuclei were co-stained using DAPI (blue). N = 4. Bar = 50 μm. (G) HUVEC cultured under static conditions were treated with varying quantities of BMP4 for 72 h. mRNA levels of HoxB9 and ID1 were quantified by qRT-PCR. N = 6 experiments. (H) HUVEC were transfected with siRNA targeting BMP4 or with scrambled control sequences and then exposed for 72 h to low WSS. mRNA levels of BMP4, HoxB9, and ID1 were quantified by qRT-PCR. N = 4 experiments. (I) HUVEC cultured under static conditions were transfected with siRNA targeting HoxB9 or with scrambled control sequences, and were treated with or without BMP4. mRNA levels of inflammatory molecules and HoxB9 were quantified by qRT-PCR. N = 6 independent experiments. (A–I) *P < 0.05, **P < 0.01, ***P < 0.001 using a paired two-tailed t-test (A and I), unpaired two-tailed t-test (D, E, and H), and one-way ANOVA (C and G).