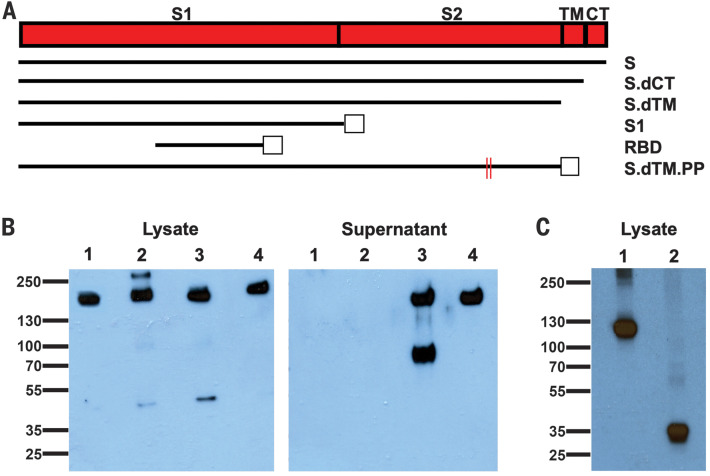
Fig. 1. Construction of candidate DNA vaccines against SARS-CoV-2.
(A) Six DNA vaccines were produced expressing different SARS-CoV-2 spike (S) variants: (i) full length (S), (ii) deletion of the cytoplasmic tail (S.dCT), (iii) deletion of the transmembrane (TM) domain and cytoplasmic tail (CT) reflecting the soluble ectodomain (S.dTM), (iv) S1 domain with a foldon trimerization tag (S1), (v) receptor-binding domain with a foldon trimerization tag (RBD), and (vi) prefusion-stabilized soluble ectodomain with deletion of the furin cleavage site, two proline mutations, and a foldon trimerization tag (S.dTM.PP). Open squares depict foldon trimerization tags; red lines depict proline mutations. (B) Western blot analyses for expression from DNA vaccines encoding S (lane 1), S.dCT (lane 2), S.dTM (lane 3), and S.dTM.PP (lane 4) in cell lysates and culture supernatants using an anti-SARS polyclonal antibody (BEI Resources). (C) Western blot analyses for expression from DNA vaccines encoding S1 (lane 1) and RBD (lane 2) in cell lysates using an anti–SARS-CoV-2 RBD polyclonal antibody (Sino Biological).