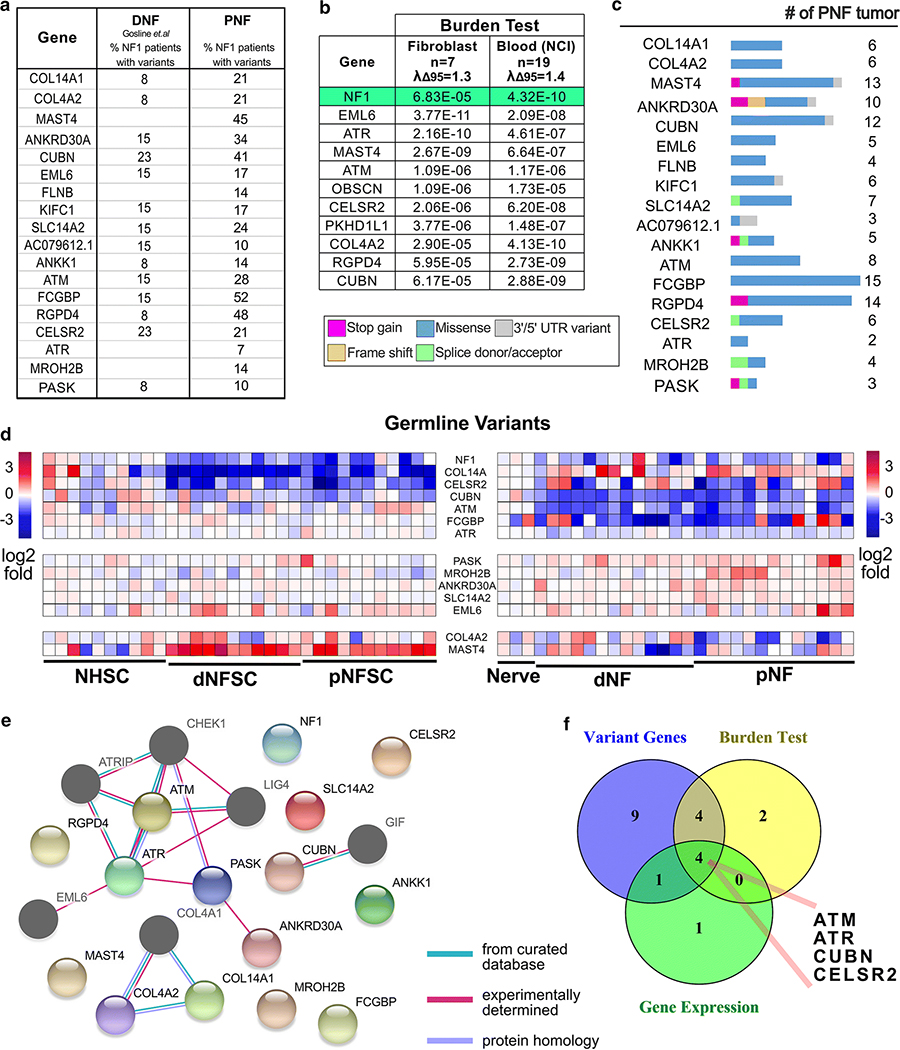
Figure 2. Genes harboring germline variants.
(a) A list of non-synonymous rare (MAF < 0.01) germline variants. (b) Genes with significant burdens (p < 1E-5) predicted by burden tests using non-common (allele frequency < 0.05) non-synonymous variants found in protein coding regions, against ExAC’s ad mixed American (AMR) sub-population. The level of artifact inflation/deflation were measured using the λΔ95 metric implemented in TRAPD. We detected the small levels of inflation (i.e. above the expected λΔ95 > 1.00) and could not eliminate entire artifacts probably due to the small case sample size and incomplete ancestry information. (c) The percent of PNF (average of cell data set from Figure 1 and Pemov et al. data) and DNF containing variants in each designated gene are shown. (d) Heatmap of RNA expression of those genes with rare germline variants in tumor SC vs. normal SC and/or tumor tissue vs. nerve. (e) STRING analysis showing the predicted interactions (confidence level >= 0.15, Maximum interaction number of the first shell <= 5) of the candidate genes, and identifies DNA damage and repair as a candidate pathway. Nodes shown in color are genes with variants identified in PNF tumors. The dark-gray nodes are genes are cross-link genes. (f) A Venny diagram showing the candidate gene filtering strategy.