FIGURE 3.
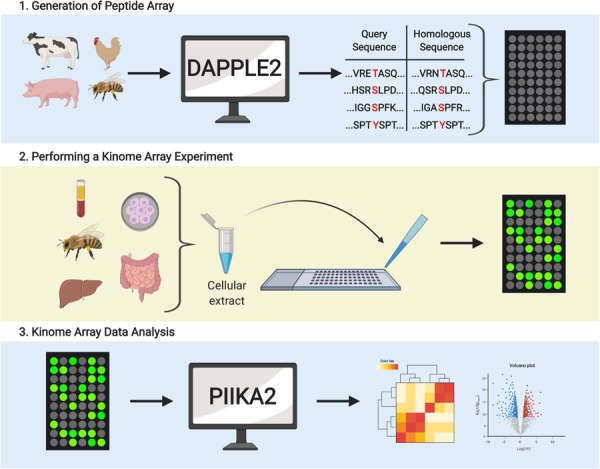
Overview of the kinome array workflow. Schematic representation of the three main components of the kinome array pipeline. (1) Computational workflow (DAPPLE2) for predicting putative phosphoacceptor peptides for spot printing on peptide microarrays. (2) Representation of the various cells, tissues and whole organisms in which cellular extracts can be prepared to quantify kinase-mediated phosphorylation using peptide arrays. (3) Computational workflow (PIIKA2) for transforming, statistical analysis and graphical visualization of kinome array data.
