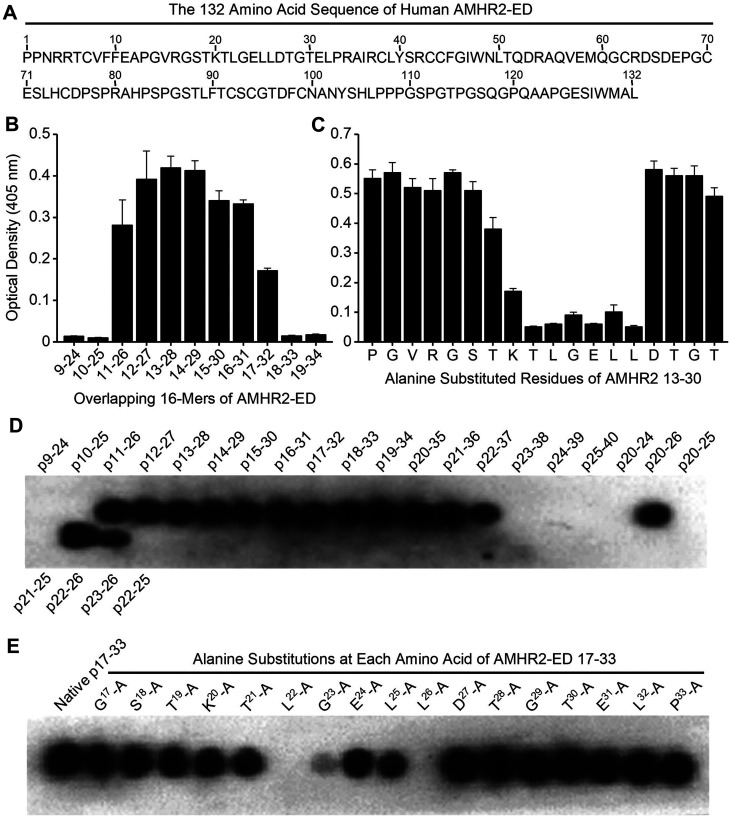
Figure 3. Identification of the AMHR2-ED binding site for the 4D12G1 mAb.
(A) The entire 132 amino acid sequence of human AMHR2-ED. (B) An overlapping series of 16-mer peptides spanning the entire sequence of human AMHR2-ED with one amino acid shifts were plated for direct ELISA testing using the 4D12G1 mAb as the primary antibody. The 4D12G1 mAb recognized residues AMHR2-ED 11–32. (C) Overlapping peptides spanning AMHR2-ED 13–30 were synthesized with alanine substitutions at each N-terminal residue or with glycine substitutions for any native N-terminal alanine residues. Competitive ELISA results showed that alanine substitutions at residues spanning AMHR2-ED 20-26 (20KTLGELL26) decreased binding of the 4D12G1 mAb to AMHR2-ED. (D) SPOT peptide arrays using 4-16-mer peptides spanning AMHR2-ED 9-40 were immobilized on cellulose membranes, treated with the 4D12G1 mAb, and the bound antibody was detected by chemiluminescence. The results showed that the AMHR2-ED 22–26 5-mer sequence (22LGELL26) represents the minimal sequence for binding of the 4D12G1 mAb. (E) SPOT peptide arrays were made using membrane bound 17-mer peptides spanning the AMHR2-ED 17–33 domain and containing alanine substitutions at each sequential amino acid. Alanine replacement of Leu22, Gly23, and Leu26 completely abolished binding by the 4D12G1 mAb. All error bars indicate ±SD, and all experiments are representative of three experiments yielding similar data.