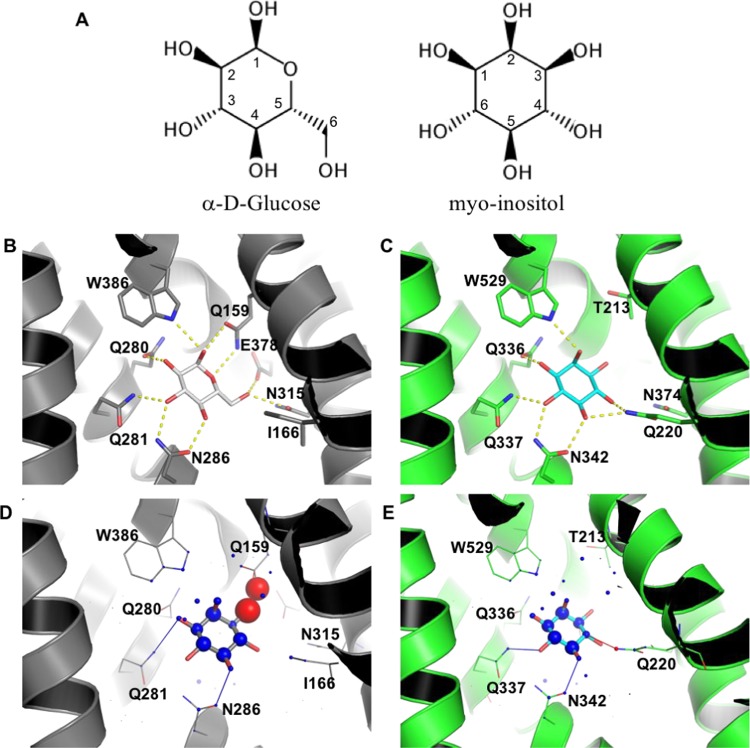
Figure 3.
Molecular bases for the specific binding of myo-inositol to GLUT13. (A) Chemical structures of α-d-glucose and myo-inositol. (B) Glucose binding mode to GLUT3 (PDB 4ZW9). (C) myo-inositol binding mode to GLUT13; prediction based on docking with PLANTS. Highlighted residues (shown as sticks) are predicted to be important for ligand binding, either by their involvement in hydrogen bonds or through improvement of sterical complementarity. Hydrogen bonds (distance cutoff = 3.5 Å) are shown as yellow dashed lines. Visualization of myo-inositol docking to GLUT3 (D) and GLUT13 (E) with DSX online. Favorable and unfavorable potentials are shown as blue and red spheres, respectively. Favorable and unfavorable distances are shown as blue and red lines, respectively.