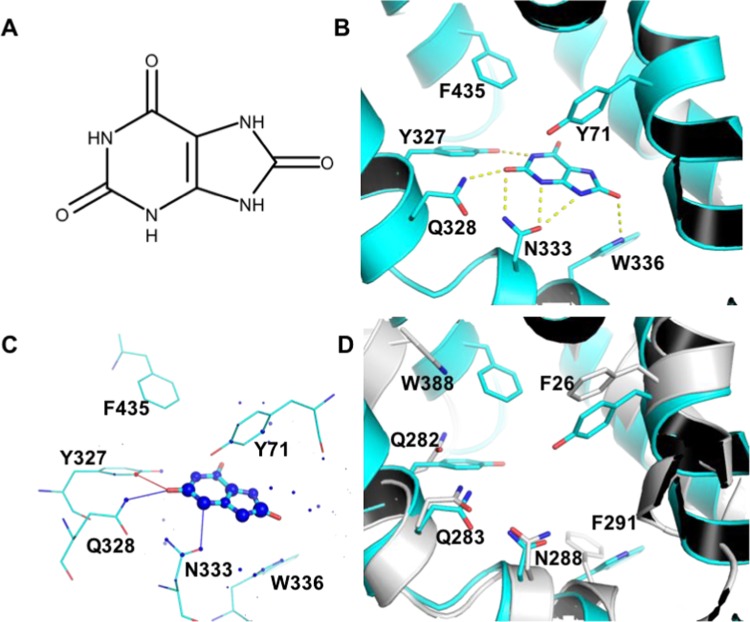
Figure 4.
Molecular bases for urate specific binding of GLUT9. (A) Urate chemical structure. (B) urate binding mode to GLUT9, predicted by docking with PLANTS. Residues predicted to hydrogen-bond with the ligand or which differ from most GLUTs are highlighted residues shown as sticks. Hydrogen bonds (distance cutoff = 3.5 Å) are shown as yellow dashed lines. (C) Visualization of the docking result with DSX online. Favorable and unfavorable potentials are shown as blue and red spheres, respectively. Favorable and unfavorable distances are shown as blue and red lines, respectively. (D) superposition of GLUT9 (cyan) and GLUT1 (white) binding sites.