Fig. 1. Resistance-associated MΦ signaling networks in MAPK-mutant tumors.
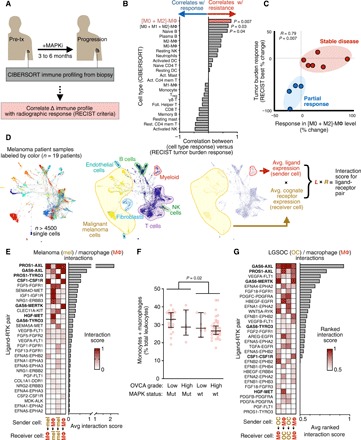
(A) Schematic depicting correlation analysis of patient biopsy immune profiling with radiographic response, used to generate data in (B) and (C). (B and C) From matched pre-MAPKi and at-progression biopsies, leukocyte change was correlated with best change in tumor burden following MAPKi in patients with melanoma (n = 9), shown across all CIBERSORT-quantified cell types (B) and with individual patient data points for the most significant immune correlate (C) (Spearman exact test with false discovery rate correction). Treg, regulatory T cells; NK, natural killer; wt, wild type; DC, dendritic cells. (D) SPRING visualization of single-cell RNA-sequencing (scRNA-seq) data from patients with melanoma, shown with individual cells pseudocolored according to the patient from which they were isolated (left) or to their annotated cell type (center). For global ligand-receptor coexpression analysis, average ligand expression levels of sender cells were multiplied with average cognate receptor expression levels of receiver cells (right). (E) Top growth factor/RTK coexpression tabulated from data in (D) and ranked according to scores between melanoma cells and MΦ (n = 19 patients). FGF, fibroblast growth factor; FGFR, fibroblast growth factor receptor. (F) Monocyte and MΦ abundance was quantified from OVCA biopsies using CIBERSORT and compared across tumors with or without RAS-MAPK–associated mutations (n = 69, medians ± interquartile range, two-tailed Mann-Whitney U test). (G) Top growth factor/RTK coexpression tabulated from LGSOC cancer cells (n = 3 patients) and ascites MΦ (n = 5 patients).
