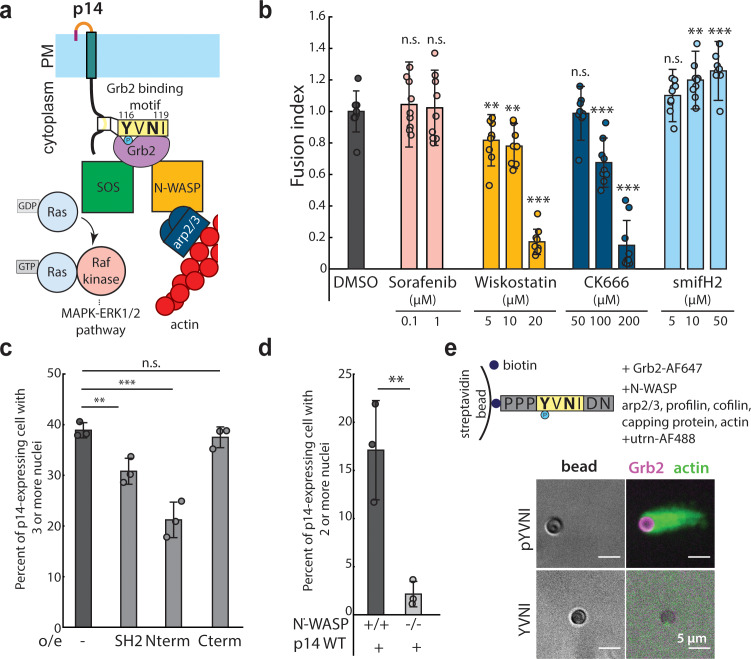
Figure 4. N-WASP-dependent assembly of branched actin network is necessary for cell-cell fusion.
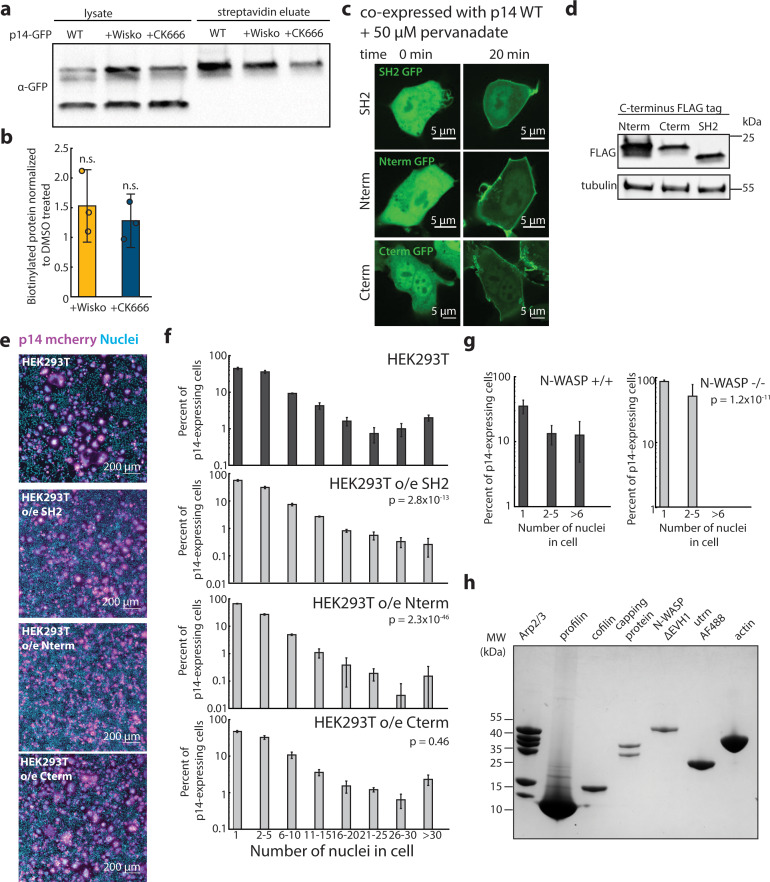
(a) Schematic of Grb2 binding to two potential downstream effectors, SOS and N-WASP (b) Extent of cell-cell fusion quantified with splitYFP fluorescence assay of p14 expressing cells treated sorafenib tosylate targeting Raf kinase, wiskostatin targeting N-WASP, CK-666 targeting Arp2/3 and smifH2 targeting formins, normalized to that of p14 WT treated with vehicle control, DMSO. Error bars indicate standard deviations from 3 independent transfections of 3 wells each. P-values are two-tailed, two-sample Student’s t-test to DMSO where ** = p<0.01, *** = p<0.001 and n.s. = p>0.05. (c) Average percent of p14-expressing cells with 3 or more nuclei in HEK293T WT cells and HEK293T cells overexpressing Grb2 SH2 domain, N-terminus SH2-SH3 mutant and C-terminus SH2-SH3 mutant. P-values are two-tailed, two-sample Student’s t-test where ** = p<0.01, *** = p<0.001 and n.s. = p>0.05. Error bars represent standard deviations from 3 independent transfections (See also Figure 4—figure supplement 1a,b,c,d, and Figure 4—source data 1). (d) Average percent of p14-expressing cells with 2 or more nuclei in N-WASP -/- and +/+ cells with error bars representing standard deviations from 3 independent transfections. P-values are two-tailed, two-sample Student’s t-test where ** = p<0.01 (See also Figure 4—figure supplement 1g). (e) In vitro actin bead motility of phosphorylated p14 cytoplasmic tail peptide conjugated to streptavidin beads in a purified actin motility mixture supplemented with Grb2. Polymerized actin is visualized with AlexaFluor488-labeled utrophin actin binding domain (See also Figure 4—figure supplement 1h).