Figure 5. Branched actin assembly directly coupled to p14 cytoplasmic tail drives cell-cell fusion.
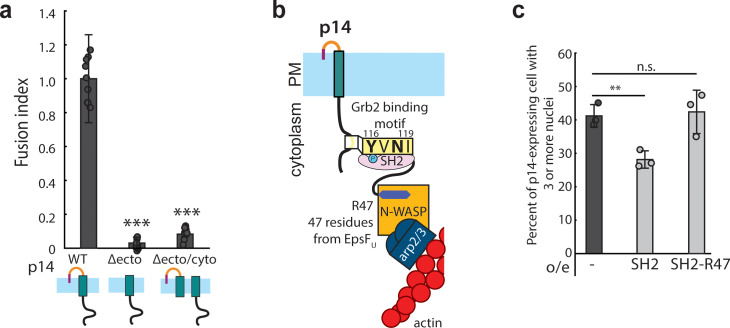
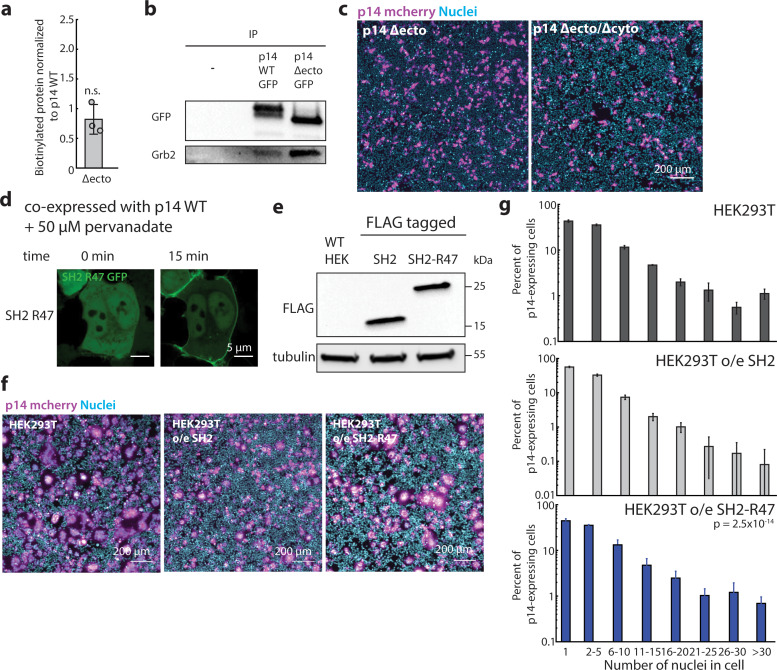
(a) Average fusion index of p14 truncation mutants normalized to that of p14 WT. P-values are two-tailed, two-sample Student’s t-test to p14 WT where *** = p<0.001. Error bars indicate standard deviations from 3 independent transfections of 3 wells each (See also Figure 5—figure supplement 1a, (b,c). (b) Schematic of fusion protein coupling actin assembly to p14 cytoplasmic tail consisting of Grb2 SH2 domain and 47 residues from EspFU. (c) Average percent of p14-expressing cell with 3 or more nuclei in HEK293T WT cells and HEK293T cells overexpressing Grb2 SH2 domain and SH2-R47. P-values are two-tailed, two-sample Student’s t-test where ** = p<0.01 and n.s. = p>0.05. Error bars represent standard deviations from 3 independent transfections (See also Figure 5—figure supplement 1d,e,f,g, and Figure 5—source data 1).