Abstract
Long noncoding RNAs (lncRNAs) regulating diverse cellular processes implicate in many diseases. However, the function of lncRNAs in cellular senescence remains largely unknown. Here we identify a novel long intergenic noncoding RNA Linc-ASEN expresses in prematurely senescent cells. We find that Linc-ASEN associates with UPF1 by RNA pulldown mass spectrometry analysis, and represses cellular senescence by reducing p21 production transcriptionally and posttranscriptionally. Mechanistically, the Linc-ASEN-UPF1 complex suppressed p21 transcription by recruiting Polycomb Repressive Complex 1 (PRC1) and PRC2 to the p21 locus, and thereby preventing binding of the transcriptional activator p53 on the p21 promoter through histone modification. In addition, the Linc-ASEN-UPF1 complex repressed p21 expression posttranscriptionally by enhancing p21 mRNA decay in association with DCP1A. Accordingly, Linc-ASEN levels were found to correlate inversely with p21 mRNA levels in tumors from patient-derived mouse xenograft, in various human cancer tissues, and in aged mice tissues. Our results reveal that Linc-ASEN prevents cellular senescence by reducing the transcription and stability of p21 mRNA in concert with UPF1, and suggest that Linc-ASEN might be a potential therapeutic target in processes influenced by senescence, including cancer.
Subject terms: Cell biology, Molecular biology
Introduction
Cellular senescence is defined as a state of indefinite growth arrest that can be induced by a range of intrinsic and extrinsic stresses including telomere shortening, oncogene activation, DNA damage, mitochondrial dysfunction, and oxidative injury [1, 2]. Cellular senescence is considered a fail-safe mechanism to prevent the propagation of severely damaged cells. Senescent cells exhibit typical characteristics, such as an enlarged and flattened morphology, genomic instability, increased lysosomal content, epigenetic alterations, and apoptosis resistance [1, 2]. Senescent cells remain metabolically active and secrete a myriad of factors that affect the neighboring cells through cell-to-cell communication [3]. Cellular senescence is triggered primarily by two complementary p53-p21 and p16-pRb pathways [4]. In the context of cancer, senescence has been proposed as a tumor-suppressive mechanism, particularly in young individuals [4–7]. Paradoxically, however, the burden of senescent cells accumulating in tissues with advancing age is believed to be protumorigenic; this influence has been linked to the excessive production of cytokines, growth factors, and matrix metalloproteases that comprise the senescence-associated secretory phenotype (SASP) [8–10]. Recent studies have found that cellular senescence is also involved in embryonic development, which has a distinct molecular pathway from senescence in nonembryonic tissues [11]. The increased interest has resulted in significant progress in cellular senescence over the last decade. Recent studies focused not only on the pathological and physiological significance, but also on the potential therapeutic application of cellular senescence.
Long noncoding RNAs (lncRNAs) are a major class of noncoding RNAs, which are defined as being longer than 200 nt transcribed by RNA polymerase II and commonly originated from intergenic regions. LncRNAs are capped, spliced, and polyadenylated. They influence the distinct cellular function including the cell fate determination, cell cycle progression, and apoptosis and involve in both physiological and disease-related process [12–14]. Many lncRNAs act directly or indirectly as oncogenes or tumor suppressors [15]. The number of lncRNA genes has been broadly estimated, ranging from <20,000 to >100,000 in humans [16]. On the other hand, the biological function and relevance of huge numbers of lncRNAs are still unknown. Emerging studies have shown that lncRNAs play important and divergent roles in the regulatory programs of gene expression at the transcriptional or posttranscriptional levels [17–19]. LncRNAs carry out these functions by interacting with other RNAs, with DNA, and with proteins in both the nucleus and cytoplasm [19, 20]. Nuclear lncRNAs control chromosome architecture and recruit transcriptional regulatory factors, including Polycomb Repressive Complex (PRC), to different chromosomal loci [21–24]. PRC1 and PRC2 place a repressive histone mark, histone 3 lysine 27 tri-methylation (H3K27me3), on the chromatin of target genes to elicit silencing of gene transcription [25, 26]. LncRNAs found in the cytoplasm enable the function of cytoplasmic organelles, assemble cytoplasmic complexes, and sequester various cytosolic regulatory factors [19, 27–29].
Senescence-associated lncRNAs have been unraveled in recent years [30, 31], but the identification and characterization of novel lncRNAs having a critical role in cellular senescence are still in demand. This study describes the identification of a novel long intergenic noncoding RNA associated with senescence (Linc-ASEN) from whole RNA-sequencing (RNA-Seq). Our results reveal that Linc-ASEN complexes with UPF1 (up-frameshift 1, also known as RENT1) and sequentially lowers the production of senescence protein p21 by repressing p21 mRNA transcription and triggering cytoplasmic p21 mRNA decay.
Results
Linc-ASEN depletion leads to cellular senescence
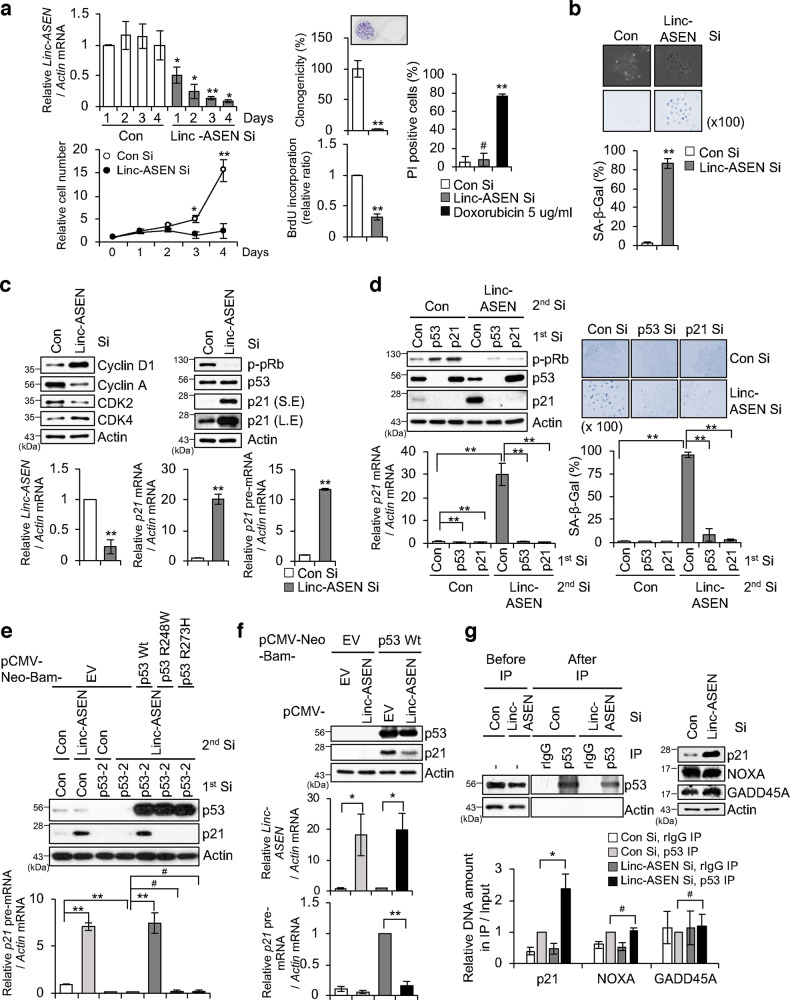
To identify novel lncRNAs related to cellular senescence, we conducted whole RNA-Seq of prematurely senescent human breast carcinoma MCF7 cells. Novel transcripts that were commonly induced in both IR- and doxorubicin-induced senescent MCF7 cells were assembled from reads aligned to human hg19 reference genome (GRCh37/UCSC hg19). Unannotated five novel lncRNAs were selected based on transcript length and protein-coding potential using the UCSC Genome Browser Database (Supplementary Table 1). To identify lncRNAs with functional roles in cellular senescence, MCF7 cells were transfected with siRNAs (Si) designed to silence each of the five selected lncRNAs, whereupon we assessed cell numbers, positivity of senescence-associated β-galactosidase (SA-β-Gal) staining (a marker of cellular senescence), and p21/p-pRb levels were monitored (Supplementary Fig. 1). We found that depletion of uncharacterized transcript #4 resulted in typical senescent traits, such as decreased cell proliferation and BrdU incorporation, flattened and enlarged cell morphology, SA-β-Gal positivity, accumulation of p21 (CDKN1A), and level changes of cell cycle regulatory proteins, which is named Linc-ASEN (Supplementary Fig. 1 and Fig. 1a–c). Furthermore, we observed that Linc-ASEN depletion-induced SASP factors and senescence-associated heterochromatin foci in MCF7 cells (Supplementary Fig. 2a, b).
Fig. 1. Loss of Linc-ASEN induces cellular senescence.
a Relative cell number, clonogenicity, BrdU incorporation, and PI positivity in Linc-ASEN silenced MCF7 cells. b SA-β-Gal activity in Linc-ASEN silenced MCF7 cells. Percentages of SA-β-Gal positive cells are plotted. c Immunoblotting and RT-qPCR analyses were performed in Linc-ASEN silenced cells. Actin was used as a loading control. d Linc-ASEN depletion-mediated cellular senescence after silencing either p53 or p21. After transfection of either p53 siRNA (Si) or p21 Si in the combination of Linc-ASEN Si, western blot, RT-qPCR, and SA-β-Gal analyses were conducted. e, f Western blot and RT-qPCR analyses were conducted after transfection of either p53-wildtype (Wt) or -mutants (Muts) in the absence of Linc-ASEN in MCF7 cells (e) by or ectopically overexpressing either p53 Wt or Linc-ASEN (f) in p53 null H1299 cells. g ChIP-qPCR showing p53 binding affinity changes in p21, NOXA, and GADD45A promoters in Linc-ASEN-depleted cells. ChIP was performed with anti-p53 antibody in Linc-ASEN Si-transfected cells. In a–g, data are represented as means ± SD. *P < 0.05, **P < 0.01, #P > 0.05 (Student’s t-test).
Analysis by RACE (5′ and 3′ rapid amplification of cDNA ends) revealed that Linc-ASEN was transcribed as a single exon of 794 nucleotides containing a putative polyadenylation signal (AAUAAA) located 25 nucleotides upstream of a poly(A) tract (Supplementary Fig. 2c). The assessment of protein-coding potential revealed that Linc-ASEN is lack of protein-coding capability (Supplementary Fig. 2d). Depletion of Linc-ASEN increased the levels of p21 pre-mRNA, p21 mRNA, and p21 protein (Fig. 1c). Linc-ASEN localized mainly in the nucleus, but it was also detected in the cytoplasm (Supplementary Fig. 2e). The specificity of silencing Linc-ASEN on the senescence phenotype was confirmed using an alternative siRNA (Supplementary Fig. 2f–h). We included a UCSC genome browser view of the Linc-ASEN locus, confirming that Linc-ASEN depletion did not affect the expression of four neighboring genes located upstream or downstream of the Linc-ASEN locus (Supplementary Fig. 3). Utilizing BLAT as an alignment tool [32], we identified a murine homolog Linc-asen with a different size (Supplementary Fig. 4a, b). In humans, the Linc-ASEN gene resides in chromosome 17; this locus is syntenic and conserved in a noncoding region in mouse chromosome 11. Moreover, Linc-asen silencing also induced senescence and p21 accumulation in mouse NIH3T3 fibroblasts (Supplementary Fig. 4c). In sum, we have identified Linc-ASEN as an lncRNA that suppresses cellular senescence in both human and mouse cells.
p53 and p21 are essential for Linc-ASEN depletion-induced cellular senescence
To elucidate functional role of Linc-ASEN in cellular senescence, we performed transcriptome analysis of Linc-ASEN-silenced MCF7 cells by RNA-Seq analysis. The altered abundance of several mRNAs in cells with silenced Linc-ASEN (Supplementary Table 2 and Supplementary Fig. 5a) was confirmed by RT-qPCR analysis (Supplementary Fig. 5b, c). Interestingly, p21 mRNA was the most highly induced transcript in Linc-ASEN-depleted MCF7 cells. We next examined whether p21 and its transcription factor p53 were required for the senescent phenotype induced by Linc-ASEN silencing. As shown, silencing p53 prevented p21 induction in cells rendered senescent by reduction of Linc-ASEN levels, and the rise in SA-β-Gal activity was prevented by silencing either p53 or p21 (Fig. 1d). Similarly, silencing Linc-ASEN in human colon carcinoma HCT116 cells lacking both alleles of p53 or p21 (p53−/−, p21−/−) failed to trigger senescence (Supplementary Fig. 6a). Consistent with these observations, ectopic expression of a p53 mutants (Muts; p53 R248W and p53 R273H) lacking DNA binding activity in p53-depleted MCF7 cells inhibited p21 pre-mRNA and p21 protein accumulation despite Linc-ASEN silencing (Fig. 1e). In addition, Linc-ASEN depletion-induced p21 accumulation and cellular senescence in human breast epithelial MCF10A cells expressing wild type (Wt) p53, while human breast carcinoma MDA-MB-231 cells expressing a Mut p53 (R280K) did not display senescence after silencing Linc-ASEN (Supplementary Fig. 6b, c). When we examined expression level changes of other senescence markers, we observed increased expressions of p16 and SASP factors, such as IL-8, IL-6, IL-1α, PAI-1, and MMP3, in HCT116 parental and MCF10A Wt p53 cells. In contrast, expressions of these genes were not increased in MDA-MB-231 Mut p53 cells (Supplementary Fig. 6a–c). Doxorubicin-induced senescent cells were used as a positive control group. The overexpression of Linc-ASEN strongly attenuated the transcriptional increase in p21 level in p53-overexpressing human lung carcinoma H1299 p53 deficient cells (Fig. 1f). Together, these results indicate that both p53 and p21 are required for the cellular senescence triggered by silencing Linc-ASEN.
We performed chromatin immunoprecipitation (ChIP) followed by qPCR analysis to determine whether Linc-ASEN influences p53 binding at the promoters of p21 and other p53 transcriptional target genes, NOXA, and GADD45A. Whereas p53 was recruited to the p21 promoter, p53 was not recruited to the NOXA or GADD45A promoters in the absence of Linc-ASEN (Fig. 1g), suggesting that Linc-ASEN specifically regulates p21 transcription by recruiting p53 to p21 DNA regulatory region.
Linc-ASEN binds PRC1/2 to control p21 transcription
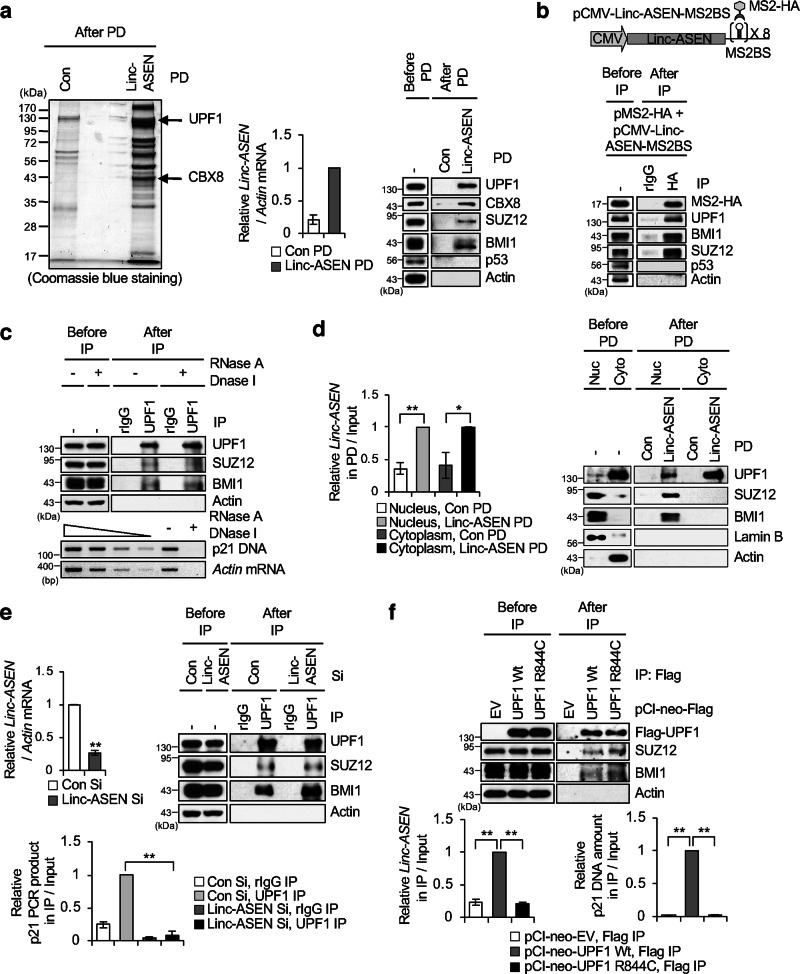
To elucidate the molecular mechanisms of Linc-ASEN action, we attempted to identify the protein partners of Linc-ASEN by adopting a modified chromatin isolation by RNA purification (ChIRP) method [33]. Proteins bound to Linc-ASEN were resolved on SDS-PAGE and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. 2a). Among the bound proteins, we found UPF1, a key regulator of nonsense-mediated mRNA decay (NMD), and CBX8, a core subunit of PRC1 (Fig. 2a). Using MS tethering system [34], we observed that UPF1, CBX8, BMI1 (a PRC1 subunit), and SUZ12 (a PRC2 subunit) were enriched in Linc-ASEN (Fig. 2b). Interactions between UPF1 and PRC1/2 subunits were not dependent on DNA or RNA (Fig. 2c). We found that Linc-ASEN formed complexes with UPF1 present in both nuclear and cytoplasmic fractions, but it formed complexes with BMI1 and SUZ12 present only in nuclear PD fractions using biotinylated Linc-ASEN probes (Fig. 2d). When we performed ChIP assay using anti-UPF1 antibody in MCF7 cells in which Linc-ASEN was silenced, UPF1 still associated with SUZ12 and BMI1, but not with the p21 gene (Fig. 2e). Moreover, the UPF1 helicase activity was dispensable for its interactions with SUZ12 or BMI1, since the helicase mutant UPF1 R844C [35] was still found to interact (Fig. 2f). In contrast, the UPF1 helicase activity was essential for UPF1 binding to the Linc-ASEN and p21 gene (Fig. 2f). UPF1 silencing alone was sufficient to induce cellular senescence, elevate p21, and increase binding of p53 at the p21 promoter, regardless of the presence of Linc-ASEN (Supplementary Fig. 7a–c). In addition, we examined the effect of Linc-ASEN or UPF1 on the enrichment of SUZ12 and BMI1 at the p21 promoter. The absence of Linc-ASEN or UPF1 significantly decreased SUZ12 and BMI1 enrichment at the p21 promoter (Supplementary Fig. 8a–d). Linc-asen also regulated p21 expression via the association with UPF1, CBX8, and SUZ12 in mouse NIH3T3 cells, indicating that the molecular mechanism whereby Linc-ASEN suppresses cellular senescence is conserved between mice and humans (Supplementary Fig. 10).
Fig. 2. Linc-ASEN associates with UPF1 and PRC1/2, and recruits UPF1-PRC1/2 complexes to the p21 gene locus.
a Linc-ASEN pulldown (PD) assay was performed and the associated proteins were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). UPF1 and PRC1/2 in Linc-ASEN RNA PD were analyzed by western blot with the indicated antibodies. b UPF1 and PRC1/2 in Linc-ASEN HA IP were analyzed by western blot with the indicated antibodies. c UPF1 associates with PRCs in a DNA/RNA-independent manner. After IP using anti-UPF1 antibody in the presence of RNase A and DNase I, the indicated proteins in precipitates were detected by western blot (upper). Actin mRNA and p21 DNA were detected to assess the extent of digestion with RNase A and DNase I (lower). d Analysis of Linc-ASEN association with UPF1 and PRCs in the nucleus and cytoplasm fractions. Lamin B and Actin were used as loading control of the nucleus and cytoplasm, respectively. e UPF1 chromatin immunoprecipitation (ChIP) was conducted in the presence or absence of Linc-ASEN. UPF1 ChIP was analyzed by western blot and RT-qPCR analyses. Enrichment of DNA was quantified relative to the amount of input. Rabbit IgG (rIgG) was used as a negative control. f ChIP analysis in the presence of UPF1 (Wt) and UPF1 R844C (Mut). After ectopic expression of UPF1 constructs in MCF7 cells, ChIP was performed using Flag antibody. Western blot and RT-qPCR analysis were performed. Enrichment of DNA was quantified relative to the amount of input. In d–f, data are represented as means ± SD. *P < 0.05, **P < 0.01 (Student’s t-test).
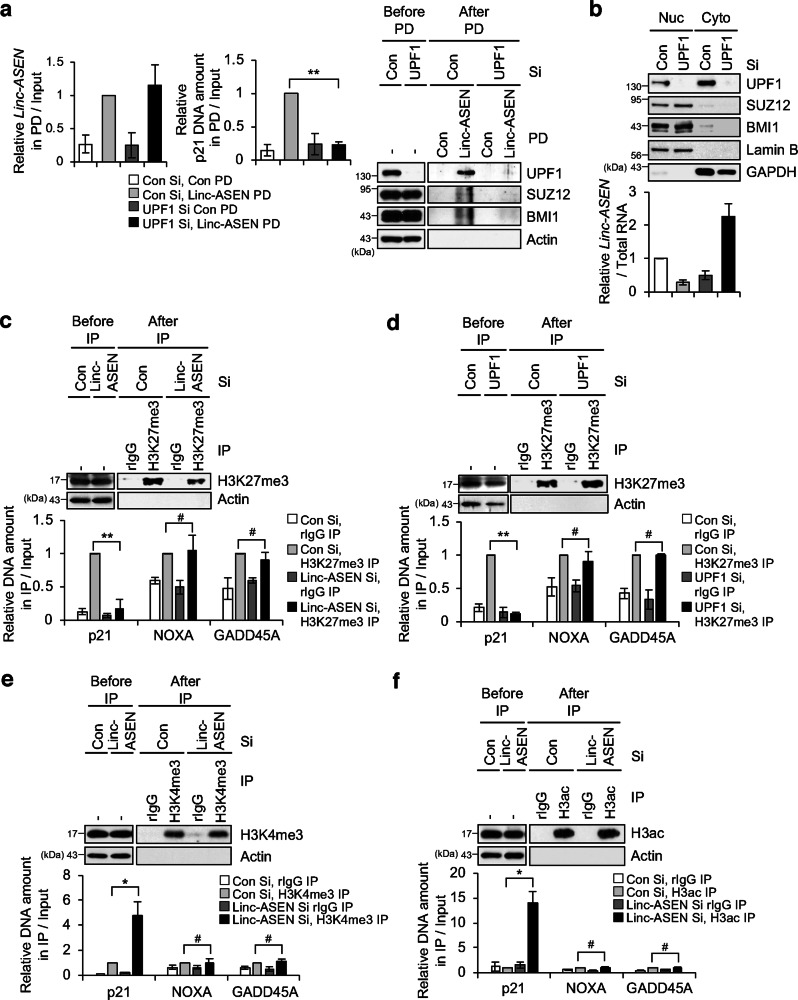
We then examined if the recruitment of PRC1/2 to the p21 gene in association with Linc-ASEN was dependent on UPF1; ChIRP analysis in UPF1-silenced cells revealed that UPF1 is essential for the recruiting PRC1 and PRC2 to the p21 gene (Fig. 3a). In addition, Linc-ASEN was primarily found in the cytoplasm in UPF1-silenced cells (Fig. 3b), suggesting that UPF1 might be needed for the nuclear localization of Linc-ASEN. Based on these findings, we hypothesized that Linc-ASEN-UPF1-PRC1/2 complexes might contribute to the epigenetic silencing of p21 gene. To test this possibility, we performed the ChIP assay for tri-methylated H3K27 (H3K27me3), a mark of transcription suppression in either Linc-ASEN- or UPF1-depleted cells. The level of H3K27me3 on the p21 promoter was substantially decreased in cells in which either Linc-ASEN or UPF1 were silenced, while it was unchanged for NOXA or GADD45A promoters (Fig. 3c, d). On the other hands, active transcription markers, H3K4me3, and acetylated histone H3 (H3ac) were higher in the p21 promoter, but NOXA and GADD45A promoters, after silencing Linc-ASEN or UPF1 (Fig. 3e, f and Supplementary Fig. 9a, b). Taken together, these results suggest that Linc-ASEN recruits PRC1/2 to the p21 gene to attenuate p21 transcription in a UPF1-dependent manner by altering the epigenetic landscape.
Fig. 3. UPF1 is required for the association between Linc-ASEN and PRCs, and is involved in histone modification of the p21 gene locus.
a ChIRP was performed using Linc-ASEN specific biotin-probes. Enrichment of DNA was quantified relative to the amount of input. b Relative Linc-ASEN levels in the nucleus and cytoplasm were assessed using nucleus and cytoplasm extraction assay after transfection of UPF1 Si. c, d H3K27me3 ChIP was performed in MCF7 cells transfected with Linc-ASEN Si (c) or UPF1 Si (d). H3K27me3 enrichment at the p21, NOXA and GADD45A promoters were determined by ChIP-qPCR analysis. Primer pairs were selected in the p53 responsive region. A rabbit IgG (rIgG) was used as a negative control. e, f ChIP was performed by H3K4me3 (e) and H3ac (f) antibodies in Linc-ASEN silenced MCF7 cells. DNA in the precipitates was measured using each specific primer and quantified relative to the amount of input. rIgG antibody was used as a negative control. In a, c–f, data are represented as means ± SD. *P < 0.05, **P < 0.01, #P > 0.05 (Student’s t-test).
ChIRP-Seq reveals specific Linc-ASEN occupancy of the p21 gene locus
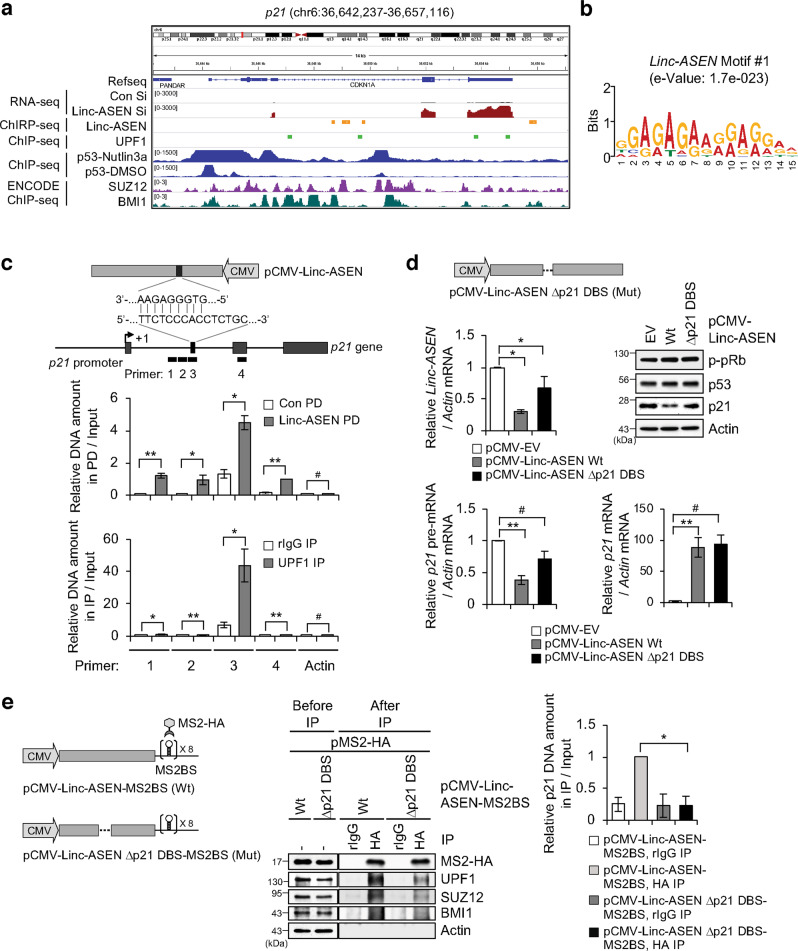
We conducted ChIRP-Seq using biotin-labeled probes of Linc-ASEN and ChIP-Seq using anti-UPF1 antibody, and identified overlaid peaks with wide genomic distribution. While there was no overlaid peak in the NOXA or GADD45A genes (Supplementary Fig. 11a), we observed an overlaid peak at p21 and ABCA12 genes, indicating Linc-ASEN-UPF1 occupancy (Fig. 4a and Supplementary Fig. 11a). Changes in the levels of p21 and ABCA12 mRNAs by either UPF1 or Linc-ASEN were confirmed using RT-qPCR analysis (Supplementary Figs. 7b and 11b). ChIRP-Seq analysis predicted that a Linc-ASEN binding motif was a purine-rich motif (e = 1.7e–023) (Fig. 4b). We validated the presence of Linc-ASEN and UPF1 in the p21 gene locus by using PCR analysis (Fig. 4c).
Fig. 4. Linc-ASEN physically binds to p21 gene locus with complementary base pairing.
a Integrated Genome Viewer (IGV 2.3) showing the expression pattern of p21 in Linc-ASEN-depleted MCF7 cells, and peaks of Linc-ASEN ChIRP-Seq and UPF1 ChIP-Seq on the p21 gene locus. b Predicted motifs enriched in Linc-ASEN peaks using MEME (http://meme-suite.org). c Schematic Linc-ASEN-binding site on the p21 gene. The Linc-ASEN and UPF1 binding sites were analyzed by ChIRP-qPCR and ChIP-qPCR, respectively. Locations of each primer (primer 1~4) are shown in the schematic. d The rate of p21 transcription in MCF7 cells transfected with pCMV-Linc-ASEN or pCMV-Linc-ASEN Δp21 DBS construct was analyzed by RT-qPCR analysis. e MCF7 cells were transfected with pCMV-Linc-ASEN-MS2BS (Wt) or pCMV-Linc-ASEN Δp21 DBS-MS2BS reporter (Mut) plasmids in combination with pMS2-HA. IP was performed using HA antibody, and then precipitated DNA was quantified using specific primers. NC, negative control. In c–e, data are represented as means ± SD. *P < 0.05, **P < 0.01, #P > 0.05 (Student’s t-test).
To explore the molecular interaction between Linc-ASEN and the p21 gene locus, we constructed a mutant plasmid that expressed a truncated transcript, Linc-ASEN Δp21 DBS, lacking the putative DNA binding sites (BS) of Linc-ASEN. As shown in Fig. 4d, while p21 expression was decreased in response to ectopic expression of Wt Linc-ASEN, Mut Linc-ASEN Δp21 DBS was not able to suppress p21 transcription. Both Wt Linc-ASEN and Mut Linc-ASEN Δp21 DBS bound to UPF1-PRC1 and UPF1-PRC2 complexes, but only Wt Linc-ASEN was capable of binding to the p21 gene locus (Fig. 4e). Taken together, these results indicate that the association of UPF1-PRC1/PRC2 complexes with Linc-ASEN is required for p21 transcriptional silencing, and suggest that Linc-ASEN leads UPF1-PRC1/PRC2 complexes selectively to the p21 gene via complementary base pairing.
Linc-ASEN cooperates with UPF1 and DCP1A to regulate p21 mRNA stability
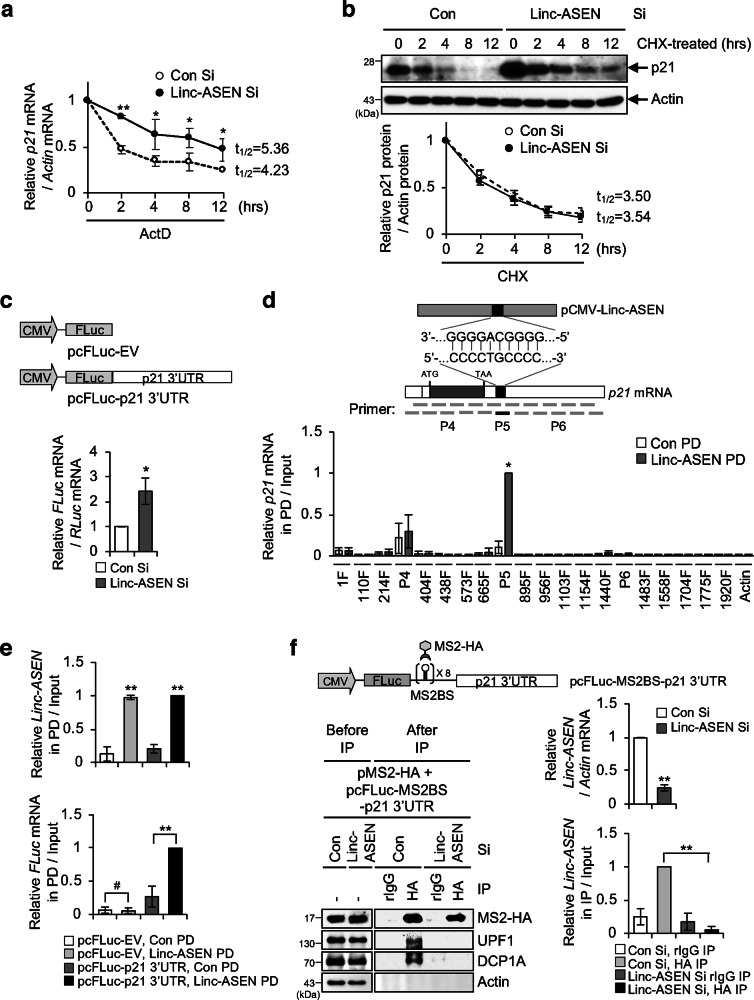
Interestingly, we found that the half-life of the p21 mRNA was significantly longer when Linc-ASEN was silenced (Fig. 5a), while p21 protein stability was not affected under these conditions (Fig. 5b). We further confirmed that the stability of p21 mRNA was controlled by Linc-ASEN using p21 3′UTR reporter construct (Fig. 5c). A putative Linc-ASEN BS on p21 mRNA was predicted by the intaRNA software (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) (Fig. 5d, top). Physical interaction between Linc-ASEN and p21 3′UTR was observed by RNA PD with biotin-labeled Linc-ASEN probe followed by RT-qPCR with p21 mRNA primers (Fig. 5d, bottom). The interaction of Linc-ASEN with p21 3′UTR was further confirmed using p21 3′UTR reporter construct (Fig. 5e). Next, we examined if UPF1 was involved in the regulation of p21 mRNA stability, since the half-life of p21 mRNA and the levels of a p21 3′UTR reporter mRNA increased following UPF1 silencing (Supplementary Fig. 12a, b). We found that Linc-ASEN and UPF1 bound to the p21 mRNA 3′UTR using RNA PD and immunoprecipitation (IP) assays, followed by RT-qPCR analysis (Fig. 5e and Supplementary Fig. 12c). UPF1 associates with multiple NMD factors, and requires decapping enzymes, such as DCP1A and DCP2, to degrade mRNA [36]. Further analysis revealed that UPF1 and DCP1A were recruited to the p21 3′UTR in a Linc-ASEN-dependent manner (Fig. 5f). However, Linc-ASEN was capable of binding to the p21 3′UTR reporter RNA regardless of UPF1 abundance, while DCP1A associated with p21 3′UTR in a UPF1-dependent manner (Supplementary Fig. 12d). Taken together, these data indicate that Linc-ASEN regulates p21 mRNA stability via the recruitment of decay molecules, such as UPF1 and DCP1A.
Fig. 5. Linc-ASEN promotes p21 mRNA decay by associating with UPF1 and DCP1A.
a Linc-ASEN silenced MCF7 cells were incubated for the indicated times with 5 mg/ml actinomycin (ActD). RNAs were purified, then subjected to RT-qPCR using p21 mRNA-specific primers. b Linc-ASEN silenced cells were treated with cycloheximide (CHX), after which lysates were prepared at the indicated times for western blot (top). The normalized band intensities were evaluated with ImageJ software (bottom). c Effect of Linc-ASEN depletion on p21 3′UTR reporter mRNA. MCF7 cells were transfected with Linc-ASEN Si prior to transfection with pFL-p21 3′UTR in combination with pRL-CMV as a reference plasmid, and then RT-qPCR analysis was conducted. d Schematic illustration showing primer binding regions of p21 mRNA for ChIRP assay (top). Predicted Linc-ASEN binding site (P5) on p21 mRNA are shown. RNA PD was performed with biotin-Linc-ASEN probes (bottom). p21 mRNA in precipitates was measured by RT-qPCR analysis with 100 nt overlapping interval primers which covers the p21 full-length mRNA. e RNA PD was performed with biotin-Linc-ASEN probes using the FLuc-p21 3′UTR reporter system. Reporter RNA in precipitates was quantified by RT-qPCR analysis using FLuc specific primers. f UPF1 binding on p21 3′UTR is dependent on Linc-ASEN. HEK293T cells were transfected with Linc-ASEN Si prior to transfection with pcFLuc-MS2BS-p21 3'UTR reporter plasmid in combination with pMS2-HA. IP was performed and immunoprecipitates were assessed by western blot and RT-qPCR analyses. In a, c–f, data are represented as means ± SD. *P < 0.05; **P < 0.01; #P > 0.05 (Student’s t-test).
Linc-ASEN binds to p21 3′UTR through complementary sequences
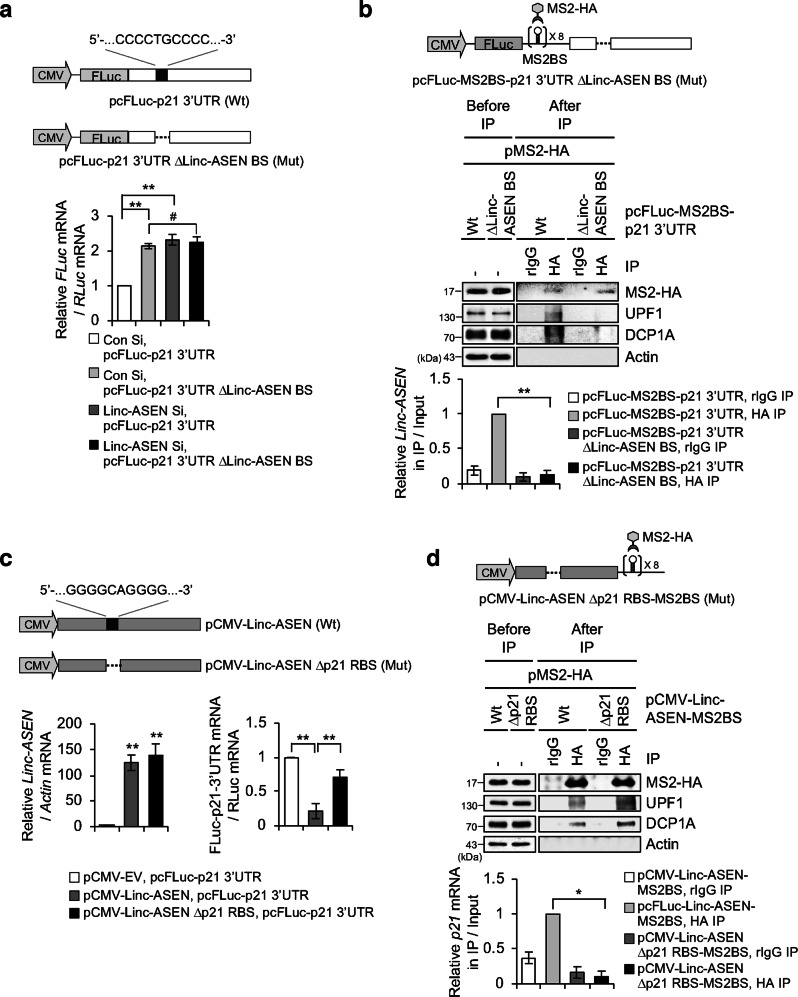
To investigate the direct binding of Linc-ASEN to the p21 3′UTR (Fig. 5d), we constructed a p21 3′UTR mutant (Mut p21 3′UTR ΔLinc-ASEN BS) in which the putative Linc-ASEN BS on p21 3′UTR was deleted (Fig. 6a). The levels of p21 3′UTR ΔLinc-ASEN BS reporter RNA were not affected by Linc-ASEN depletion (Fig. 6a), supporting the notion that the Linc-ASEN BS in the p21 3′UTR is essential for the regulation of p21 mRNA stability by Linc-ASEN. To confirm the interaction between Linc-ASEN and Linc-ASEN BS of p21 3′UTR, we constructed plasmids that expressed p21 3′UTR and p21 3′UTR ΔLinc-ASEN BS bearing MS2 hairpins. The assessment of MS2 IP materials revealed that decay-promoting molecules UPF1 and DCP1A associated with p21 3′UTR but not with p21 3′UTR ΔLinc-ASEN BS RNA (Fig. 6b). These results support the notion that Linc-ASEN BS of p21 3′UTR is essential for Linc-ASEN binding to regulate p21 mRNA stability through recruiting UPF1 and DCP1A.
Fig. 6. Linc-ASEN binding to p21 mRNA through complementary base pairing is critical for p21 mRNA degradation.
a Schematic representation of the pcFluc-p21 3′UTR (Wt) or pcFluc-p21 3′UTR ΔLinc-ASEN BS (Mut) (upper). MCF7 cells were transfected with pcFluc-p21 3′UTR Wt or Mut in combination with pRL-CMV as a reference plasmid, and then RT-qPCR analysis was performed (lower). b Diagrams of pcFLuc-MS2BS-p21 3′UTR (Wt) or pcFLuc-MS2BS-p21 3′UTR ΔLinc-ASEN BS (Mut) (upper). Using the MS2 tethering system, IP was performed, and precipitates were analyzed by western blot and RT-qPCR (lower). c Schematic representations of the pCMV-Linc-ASEN (Wt) or pCMV-Linc-ASEN Δp21 RBS (Mut) (upper). Relative Linc-ASEN and p21 mRNA levels were analyzed by RT-qPCR (lower). d MCF7 cells were transfected with pCMV-Linc-ASEN-MS2BS (Wt) or pCMV-Linc-ASEN Δp21 RBS-MS2BS (Mut) reporter plasmids in combination with pMS2-HA. Cells were subjected to IP and western blot analyses. Precipitated RNA was subjected to RT-qPCR. In a–d, data are represented as means ± SD. *P < 0.05; **P < 0.01; #P > 0.05 (Student’s t-test).
To further examine the direct binding between Linc-ASEN and p21 mRNA, we constructed Linc-ASEN mutant, which disrupted the putative p21 mRNA BS of Linc-ASEN (Linc-ASEN Δp21 RBS). Ectopic expression of Linc-ASEN Δp21 RBS did not affect p21 mRNA abundance as much as Wt Linc-ASEN overexpression did (Fig. 6c). Tethering constructs expression in transfected cells further supported the notion that p21 mRNA interacted only with Wt Linc-ASEN and not with Mut Linc-ASEN Δp21 RBS (Fig. 6d). However, UPF1 and DCP1A still associated with Linc-ASEN Δp21 RBS (Fig. 6d). Taken together, these data demonstrated that Linc-ASEN modulates p21 mRNA stability via direct interaction between Linc-ASEN (5′-GGGGCAGGGG-3′) and p21 mRNA 3′UTR (5′-CCCCTGCCCC-3′).
Linc-ASEN levels correlate closely with tumor growth and ageing
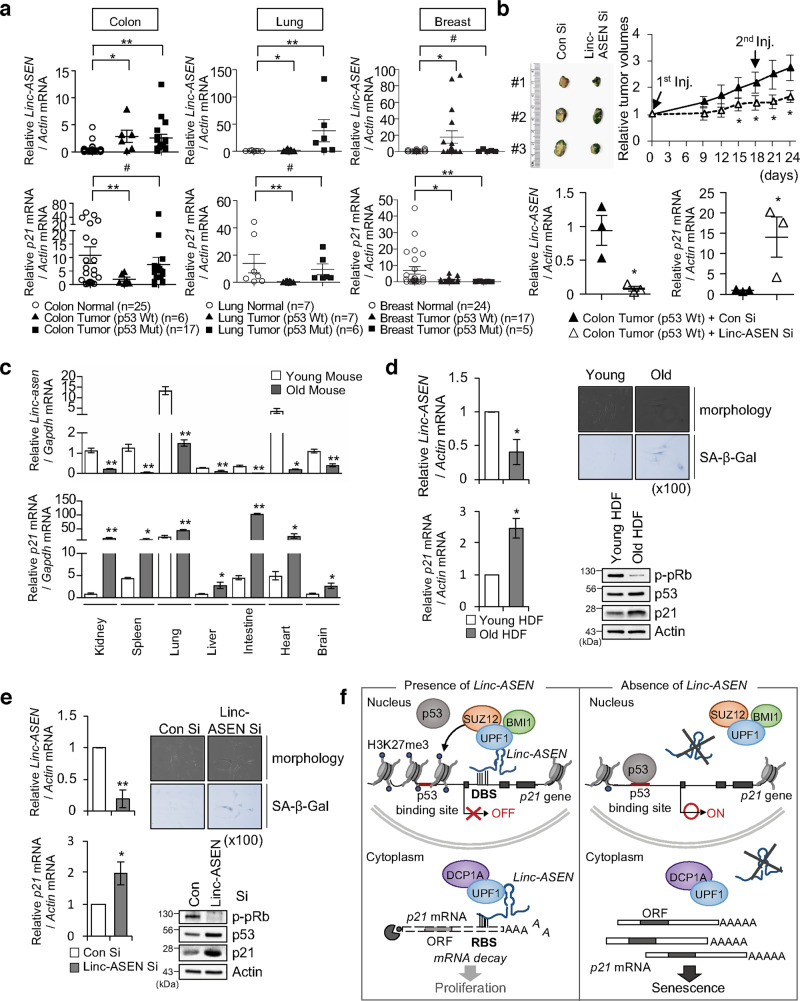
To investigate the biological significance of Linc-ASEN, we examined the correlation between Linc-ASEN and p21 mRNA levels in cancer patient tissues. Linc-ASEN and p21 mRNA levels in tumor tissues from patients with colon cancer (normal = 25, tumor = 23), lung cancer (normal = 7, tumor = 13), and breast cancer (normal = 24, tumor = 22) with normal tissues were measured by RT-qPCR analysis (Fig. 7a). The correlation between Linc-ASEN and p21 mRNA levels in cancers with Wt p53 were assessed using Pearson’s correlation coefficient. While a negative correlation was observed between Linc-ASEN and p21 mRNA expression in colon, breast, and lung cancer patients with Wt p53 (colon cancer, R = −0.576; lung cancer, R = −0.171; breast cancer, R = −0.035), no such correlation was observed in patients with Mut p53. To further investigate the biological significance of Linc-ASEN, we generated the patient-derived xenograft (PDX) mice from human colon cancer tissues with Wt p53. Significant retardation of tumor growth and increased SA-β-Gal activity were observed in a PDX mice treated with Linc-ASEN Si (Fig. 7b) and RT-qPCR analysis revealed higher p21 mRNA levels in Linc-ASEN Si treatment goup (Fig. 7b).
Fig. 7. Inverse correlation between Linc-ASEN and p21 mRNA levels in human cancer patient tissues, PDX tumor tissues, and aged mouse tissues.
a Analysis of Linc-ASEN and p21 mRNA levels in colon (normal = 25, tumor = 23), lung (normal = 7, tumor = 13) and breast (normal = 24, tumor = 22) patient samples using RT-qPCR. Expression levels were normalized to Actin mRNA. b Tumor growth curves of PDX mice (n = 3) that were generated from human colon cancer patient tumor tissues after injection of Linc-ASEN Si. Tumor volume in PDX mice was measured at the indicated times. After mice were sacrificed, tumor tissues were obtained for SA-β-Gal staining and RT-qPCR analysis. c RT-qPCR analysis of the levels of Linc-asen and p21 mRNA in young and old mouse tissues (n = 3). d The levels of Linc-ASEN and p21 mRNA were measured, and SA-β-Gal positivity evaluated, in primary human diploid fibroblasts (HDFs) that were either proliferating (passage 15) or senescent (passage 30). e Western blot and RT-qPCR analysis were performed 2 days after Linc-ASEN Si transfection in primary HDFs (passage 14). SA-β-Gal staining was performed at 4 days post transfection. f Proposed working model of Linc-ASEN. Linc-ASEN plays dual roles in inhibiting p21 expression by both suppressing transcription of the p21 gene in the nucleus and reducing p21 mRNA stability in the cytoplasm. In a–e, data are represented as means ± SEM. *P < 0.05; **P < 0.01; #P > 0.05 (Student’s t-test).
To determine if Linc-ASEN and p21 mRNA levels are correlated with aging, we investigated the relative levels of Linc-ASEN and p21 mRNA in various organs from young (3 months old) and old (24 months old) mice. With advancing age, the levels of Linc-ASEN decreased significantly, while the levels of p21 mRNA increased (Fig. 7c). Similarly, in human diploid fibroblasts (HDFs) rendered senescent by replicative exhaustion, Linc-ASEN levels were lower and p21 mRNA levels were higher (Fig. 7d) than in proliferating HDFs. We also observed the typical senescent cell morphology (flattened and enlarged) as well as positive SA-β-Gal activity in Linc-ASEN-silenced HDFs (Fig. 7e). Taken together, these data support the view that Linc-ASEN plays a critical role in tumor growth and its abundance is inversely correlated with p21 levels in untransformed senescent cells and aged tissues.
Discussion
Our studies demonstrate that Linc-ASEN functions through association with UPF1, a well-known key factor in NMD [37, 38]. UPF1 is a DNA and RNA helicase belonging to helicase superfamily 1 [39] found in both the cytoplasm and the nucleus [40]. In cytoplasm, UPF1 is critical for NMD, a conserved mRNA surveillance pathway that degrades abnormal mRNA containing a premature translation termination codon [41, 42]. In addition to its relationship with NMD, UPF1 plays roles of non-NMD, including STAU1-mediated mRNA decay (SMD), telomere maintenance, cell cycle progression, and DNA replication [28, 43, 44]. However, the function and biological significance of UPF1 in transcriptional regulation has not been investigated. Based on the results of this study, we propose that UPF1 is involved in chromatin modification via association with PRC1 and PRC2 in the nucleus. Linc-ASEN played a critical role in guiding the UPF1-PRC1 and UPF1-PRC2 complexes to the p21 gene locus through specific complementary sequences, ultimately influencing p21 gene transcription. We found that Linc-ASEN depletion led to the dissociation of UPF1-PRC1 and UPF1-PRC2 complexes from p21 gene locus, resulting in decreased H3K27me3 enrichment at a p53-responsive promoter element in the p21 gene. Accordingly, loss of Linc-ASEN contributed to derepressing the p21 promoter. By employing ChIRP-Seq and RNA IP analyses, we also reveal that loss of UPF1 caused defects in the recruitment of PRC1 and PRC2 to the Linc-ASEN binding element in the p21 gene locus.
The function of UPF in concert with lncRNA to regulate mRNA stability outside of the NMD or SMD mechanisms was not reported previously. We found that Linc-ASEN regulated p21 mRNA stability in association with UPF1 and DCP1A in the cytoplasm. The fact that Linc-ASEN contains a region complementary to the p21 3′UTR leads us to propose that Linc-ASEN could guide Linc-ASEN-UPF1-DCP1A complex to p21 3′UTR. As evidenced by studying p21 3′UTR and Linc-ASEN RNAs bearing internal mutations in either DNA- or RNA-binding regions, target site recognition by Linc-ASEN occurred through specific base pairing.
The regulatory networks controlling cancer development have been richly expanded through the regulation of protein-coding gene expressions by lncRNAs [15, 45, 46]. Through their impact on protein expression patterns, lncRNAs can act as oncogenes or tumor suppressors [47]. Given that cellular senescence, particularly in young individuals, is believed to represent a potent suppressor mechanism, we assessed the consequences of reducing Linc-ASEN levels on tumor growth in a PDX mouse model of human colon cancer bearing Wt or Mut p53. Depletion of Linc-ASEN reduced tumor size and triggered a strong response of tumor cell senescence. In addition, we found a negative correlation between Linc-ASEN levels and p21 mRNA levels in patient cancers containing Wt p53, and discovered that Linc-ASEN levels decreased as p21 levels increased in aged mouse tissues.
In summary, we report a novel transcript, Linc-ASEN, with a dual function in fine-tuning p21 production at both the transcriptional and posttranscriptional levels (Fig. 7f). In the nucleus, Linc-ASEN guided UPF1-PRC1 and UPF1-PRC2 to the p21 gene promote to suppress p21 gene transcription. In the cytoplasm, Linc-ASEN led the UPF1-DCP1A complex to the p21 3′UTR in a sequence-specific manner to degrade p21 mRNA. Our data indicate that the presence of Linc-ASEN robustly prevents cellular senescence. Given the role of Linc-ASEN as a strong, multileveled inhibitor of p21 expression, we propose that interventions to reduce Linc-ASEN function could constitute promising therapeutic venues in cancer.
Materials and methods
Cell culture
MCF7, HEK293T, and SW620 cells and HDFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (WelGENE, Daegu, Korea). HCT116 parental, p53−/−, and p21−/− isogenic cell lines were cultured in McCoy’s 5A medium (WelGENE). MDA-MB-231 and H1299 cell lines were cultured in RPMI 1640 (WelGENE). MCA10A cells were cultured in DMEM/F12 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 5% horse serum (Sigma-Aldrich, St. Louis, MO, USA) and 1% penicillin and streptomycin (WelGENE). The other cells were supplemented with 10% fetal bovine serum (Tissue Culture Biologics, Long Beach, CA, USA) and 1% penicillin and streptomycin (WelGENE) at 37 °C in a 5% CO2 incubator.
siRNA, ASO, and plasmid transfection
Cells were transfected with in vitro-synthesized siRNAs (Bioneer, Daejeon, Korea), ASOs (Bioneer), and plasmids using Lipofectamine RNAi-MAX (Invitrogen, Carlsbad, CA, USA) and Lipofectamine 2000 (Invitrogen). The siRNAs and ASOs used in this study are listed in Supplementary Table 3.
Cell treatments
Doxorubicin was used to induce senescence or apoptosis at concentrations of 50 ng/ml or 5 µg/ml, respectively. For analyses of p21 mRNA stability and p21 protein half-life, cells were incubated with 5 µg/ml actinomycin D (Sigma-Aldrich) and 40 µg/ml cycloheximide (Sigma-Aldrich) for various time periods (0, 2, 4, 8, and 12 h). Total RNA was isolated and RT-qPCR analysis was used to measure RNA levels; protein lysates were prepared for western blot analysis of specific proteins.
Assays of cell viability, colony formation, and BrdU incorporation
Suspended cells were stained with 0.4% (w/v) trypan blue (Thermo Fisher Scientific) at a 1:1 ratio to assess cell viability, and then counted using a hemocytometer. For colony formation assay, cells were seeded in 60-mm dishes and cultured for 7 days. Colonies were then fixed and stained with Diff-Quick (Sysmex Corp., Kobe, Japan). For BrdU incorporation assay, cells were seeded into 12-well plates and cultured for 2 days. A Cell Proliferation ELISA, BrdU (colorimetric) kit (Roche, Darmstadt, Germany) was used according to the manufacturer’s instructions.
Senescence-associated β-galactosidase (SA-β-Gal) staining
Cells and tumors were stained for SA-β-Gal activity as described previously [48]. Briefly, cells were washed with phosphate-buffered saline (PBS) and fixed with 3.7% formaldehyde for 3–5 min. And then cells and tissues were incubated for 12–16 h in 1 ml staining solution containing 1 mg/ml 5-bromo-4-chloro-3-indolyl β-D-galactoside (X-Gal), 40 mM citric acid/sodium phosphate (pH 6.0), 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2.
Immunofluorescence
Cells were washed with PBS and fixed with 4% paraformaldehyde (Sigma-Aldrich) in PBS for 10 min at room temperature. And then cells were permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) and were blocked for 20 min with 1% BSA in PBS. Cells were stained with primary antibody in 1% BSA in PBS and were stained with Alex Flour 594-conjugated secondary antibody (Jackson Immunoresearch, West Grove, PA, USA). Nuclei were stained with DAPI after the incubation of secondary antibody. Next, slide glass was mounted using Fluorescent Mounting Medium (Dako, Santa Clara, CA, USA). Immunofluorescence was visualized using laser scanning microscope (LSM 510; Carl Zeiss, Jena, Germany). Antibody is listed in Supplementary Table 8.
Plasmid construction
Construction details for pMS2-HA were previously reported [49]. To generate pcFLuc-EV plasmids, pcDNA3 (Invitrogen) was ligated with a HindIII/XbaI-digested PCR fragment amplified from pGL2-basic (Promega). To construct pcFLuc-p21 3′UTR reporter plasmid, pcFLuc-EV plasmid was inserted with XbaI-digested PCR products amplified from p21 3′UTR. To generate pcFLuc-MS2BS-p21 3′UTR, NotI/XbaI restriction enzyme sites were inserted into pcFLuc-p21 3′UTR using specific primers. NotI/XbaI fragments of pcFLuc-MS2BS were then ligated with NotI/NheI-digested pcFLuc-p21 3′UTR-NotI/XbaI plasmid. pCMV-Linc-ASEN was generated from ligation of a BglII/HindIII-digested PCR fragment amplified from CMV promoter and a HindIII/XbaI-digested fragment amplified from BAC clone (Macrogen, Seoul, Korea) into BglII/XbaI-digested pcDNA3 plasmid. To construct pCMV-Linc-ASEN-MS2BS, NotI and NheI restriction enzyme sites were added into pCMV-Linc-ASEN using the primers, after which a NotI/XbaI-digested fragment from pcFLuc-MS2BS was inserted into NotI/NheI-digested pCMV-Linc-ASEN-NotI/NheI. To generate pCMV-ORF 1-GFP, -ORF 2-GFP, and -ORF 3-GFP, pCMV-Linc-ASEN was amplified using the primer pair after adding the NheI/NotI restriction enzyme site into ORF 1, ORF 2, and ORF 3 of Linc-ASEN. Next, NheI/NotI-digested pEGFP-C1 (Clontech Laboratories, Mountain View, CA, USA) was inserted into pCMV-ORF 1, 2, or 3-NheI/NotI plasmids, respectively. pCMV-Linc-ASEN Δp21 DBS, Δp21 RBS, Δp21 DBS-MS2BS or Δp21 RBS-MS2BS plasmids were constructed by ligation of HindIII/XbaI-digested PCR fragments amplified from pCMV-Linc-ASEN or pCMV-Linc-ASEN-MS2BS, respectively. To generate pcFLuc-p21 3′UTR ΔLinc-ASEN BS or -MS2BS-p21 3′UTR ΔLinc-ASEN BS, a HindIII/XhoI fragment of pcFLuc-p21 3′UTR or -MS2BS-p21 3′UTR was ligated with a HindIII/XhoI-digested PCR fragment amplified from pcFLuc-p21 3′UTR. p3xFLAG-p21 was constructed by ligation of HindIII/EcoRI-digested fragment using gBlock (IDT Corp., Coralville, IA, USA) into p3xFLAG plasmid.
The pCMV-Neo-Bam-p53 Wt, R248W, or R273H expressing plasmids were purchased from Addgene (http://www.addgene.org/). pCI-neo-EV, -FLAG-UPF1 Wt, and -FLAG-UPF1 R844C plasmids were kindly provided by Dr Lynne E. Maquat at the University of Rochester, Rochester, NY, USA [35]. The primers used for the generation of plasmid constructs are listed in Supplementary Table 4.
Subcellular fractionation
Nuclear/cytoplasmic fractionation was conducted using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) according to the manufacturer’s protocols. Cytoplasmic and nuclear fractions were split for RNA extraction for RT-qPCR analysis or protein extraction for western blot analysis. GAPDH, Actin, and HSP 90 were used as cytoplasmic protein markers and Lamin B1 was used as a nuclear protein marker in western blot analyses.
Rapid amplification of cDNA ends (RACE)
Total RNAs were extracted using TRIzol Reagent (Molecular Research Center, Cincinnati, OH, USA) following the manufacturer’s protocol, and then used for RACE. Specifically, 5′ and 3′ RACE reactions were performed using 5′ and 3′ RACE Systems for RACE, version 2.0 (Thermo Fisher Scientific), according to the manufacturer’s protocol. The sequences of primers used for RACE are listed in Supplementary Table 5.
Real-time quantitative PCR (RT-qPCR)
Total RNAs were extracted from cells or normal and cancer tissues of human colon, lung, or breast using TRIzol Reagent (Molecular Research Center), and then reverse-transcribed using M-MLV reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer’s protocols; qPCR analysis was then performed using iQTM SYBR® Green Supermix (BioRad Laboratories, Hercules, CA, USA) on a CFX ConnectTM RT-PCR Detection System (BioRad Laboratories). The relative expression levels were normalized to Actin mRNA. The primer sequences for RT-qPCR are listed in Supplementary Table 6.
RNA-pulldown assay
Harvested cells were rinsed and sonicated in NET-2 buffer (50 mM Tris-HCl, pH 7.4, 150–300 mM NaCl, 0.05% NP-40, PMSF, Benzamidine). Cell lysates were incubated with biotin-labeled probes synthesized from Bioneer, and then pulled down with streptavidin beads (Sigma-Aldrich). Precipitated RNA and proteins were subsequently subjected to RT-qPCR and western blot analyses, respectively. The sequences of biotin-labeled probes for the Linc-ASEN pulldown assay are listed in Supplementary Table 7.
RNA-Seq analysis
RNA-Seq was conducted to identify novel lncRNAs differentially expressed in senescent cells and to analyze genes that were differentially expressed in response to Linc-ASEN depletion. For each experiment, MCF7 cells were untreated or treated with DMSO, doxorubicin (50 ng/ml), or IR (6 Gy) for 24 h, and then transfected with Control or Linc-ASEN siRNAs for 2 days, respectively. Total RNA was isolated using TRIzol Reagent (Molecular Research Center) according to the manufacturer’s protocols. The cDNA was then synthesized from total RNA as previously described [50]. For high-throughput sequencing, standard Illumina libraries were prepared according to the manufacturer’s protocol and sequenced using Illumina HiSeq2000 or NextSeq 500 of read length 101 or 75 bp, respectively. We obtained ~96 million mapped reads per sample and aligned then to the human reference genome hg19 of UCSC genome using STAR v2.5.0 [51]. Differentially expressed transcripts between the controls and samples were investigated using Cufflinks v2.2.1 [52]. All expression values were calculated as FPKM (Fragments Per Kilobase Million).
Immunoprecipitation (IP) and chromatin immunoprecipitation (ChIP)
For IP and ChIP, cells were lysed in NET-2 buffer containing 100 U of RNase Inhibitor (Invitrogen) and precleared with protein G-Resin (GenScript, Piscataway, NJ, USA) or A-Sepharose beads (GE Healthcare Bio-Science AB, Uppsala, Sweden). After precleaning, the lysates were immunoprecipitated with specific antibodies or IgG along with beads. The DNAs and RNAs pulled down with proteins were purified by phenol-chloroform extraction and precipitated in ethanol, after which they were resuspended in DNase- and RNase-free water and measured by qPCR analysis. The proteins were analyzed by western blot analysis. The antibodies used in IP and ChIP are listed in the Supplementary Table 8.
Chromatin isolation by RNA purification (ChIRP) assay and data analysis
ChIRP assays were performed as previously described [33]. Briefly, three 3′ biotinylated probes targeting the full-length of the Linc-ASEN sequence were designed (see Supplementary Table 7 for details). MCF7 cells were then sonicated in NET-2 buffer and precleared with streptavidin beads (Sigma-Aldrich) at 4 °C for 1 h with end-over-end rotation. After preclearing, 2 μl per 100 pmol of each probe were added and incubated at 4 °C for 2 h. Beads:biotin-probes:RNA:chromatin were captured with streptavidin beads (Sigma-Aldrich) and resuspended in 100 μl of 2× Laemmli sample buffer and boiled for 5 min, after which they were subjected to trizol:chloroform extraction. The extracted RNA was then processed using an Illumina TruSeq Stranded mRNA kit (Illumina) according to the manufacturer’s protocol and sequenced on thane Illumina NextSeq 500 (Illumina) system with a sequence length of 75 bp. Analysis of raw data was conducted as described for the RNA-Seq data above. Peaks of each sample were called using MACS against its corresponding input with a p value < 0.01. Sequences of peaks within genes that were upregulated by Linc-ASEN were extracted and motifs analysis against these peaks was performed by MEME as described [53]. The motifs showing the highest significance were used for further analysis.
Liquid chromatography-mass spectrometry (LC-MS/MS)
MCF7 cell lysates were pulled down with streptavidin beads (Sigma-Aldrich). Briefly, precipitated proteins were stained and measured by MS via LC-MS/MS on a Q Exactive Plus mass spectrometer (Thermo Scientific) equipped with a Dionex Ultimate 3000 UPLC Orbitrap ion source (Korea Basic Science Institute, Daejeon, Korea). Spectral data were analyzed using the MASCOT Ver2.4, a database containing the human International Protein Index protein sequence database, (http://www.ebi.ac.uk/IPI/) together with the reversed complement.
Western blot analysis and antibodies
Cells were lysed with RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitor (Roche). Lysates were mixed with 2× sample buffer, boiled, and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After transferring proteins to a nitrocellulose membrane, western blot analysis was conducted by blocking the membrane with 5% nonfat dried milk and incubating with specific antibodies. The antibodies used in western blotting are listed in Supplementary Table 8.
Patient-derived xenograft (PDX)
Tumor samples were obtained from patients following surgical resection for colon cancer at Severance Hospital, Yonsei University Health System, Seoul, Republic of Korea. Samples were procured from surgical specimens and processed for subsequent analyses within a few hours. Pathological and genetic information were annotated and managed in a prospective database. All patients provided informed consent and the study was approved by the Severance Hospital Institutional Review Board (4-2013-0635).
For PDX model, 5-week-old BALB/c nude female mice were cared for according to the institutional guidelines for animal care and every animal experiment was approved by the Institutional Animal Care and Use Committee (IACUC) of the Yonsei University College of Medicine (2013-0262). The transferred tumors were placed into sterile Petri dishes containing PBS, and then sliced into fragments. Initially, a single piece of tumor tissue was used for implantation. Thereafter, when the tumor reached an average volume of ~200 mm3, 100 nM of Con Si or Linc-ASEN Si with AteloGenes (KOKEN, Tokyo, Japan) was injected to wrap the entire tumor mass. The size of the implanted tumor was checked periodically for 24 days using a caliper. Average tumor volume was calculated as (length × width2)/2. The mice were sacrificed at 24 days after siRNA injection, after which tumors obtained from mice were used for RT-qPCR and western blot analysis. The randomization method was not used and no blinding was done. The use of animals in this study was approved by IACUC of the Yonsei University College of Medicine (2013-0262).
Human tissues
Human breast normal and cancer tissues were provided by the Biobank of Korea University Guro Hospital, a member of the Korea Biobank Network. Human colorectal normal and carcinoma tissues were provided by the Ajou University Human Bio-Resource Bank, a member of Korea Biobank Network. The human tissues used in this study were approved by the Institutional Review Board of Inha University Hospital (2016-11-008).
Public data sets sources
Data for ChIP-seq of p53 at the p21 promoter were derived from a publicly available dataset (GSE86222), while those for PRC1 and PRC2 were obtained from ENCODE (BMI1 ChIP-seq, GSE105933 and SUZ12 ChIP-seq, GSE105981).
Statistical analysis
All data represent the mean ± standard deviation and standard error of the mean for at least three independent experiments. Sample numbers (n) indicates the number of independent biological samples in each experiment. Statistical analysis was carried out using Microsoft Excel software and GraphPad Prism. Statistical significance was analyzed using Student’s t test and expressed as a P value (*P < 0.05; **P < 0.01; ***P < 0.001; #P > 0.05) as indicated in the individual figure legends.
Accession numbers
Sequencing data from this paper have been submitted to the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and assigned the accession number GSE128398.
Supplementary information
Acknowledgements
We are grateful to Prof. Lynne E. Maquat at University of Rochester for pCI-neo-EV, pCI-FLAG-UPF1-WT, and pCI-FLAG-UPF1 R844C plasmids, and Dr Keetae Kim at Daegu Gyeongbuk Institute of Science and Technology (DGIST) for tissues from young and old mouse. This work was supported by grants to JSL from MRC (2014009392), Nuclear Research and Development program (2017M2A2A7A01070591), and Basic Research Science Program (2017R1A2B2007542) through the National Research Foundation (NRF) from Korean government (MSIT).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The biospecimens used for this study were provided by the Biobank of Inha University Hospital, and the Ajou University Human Bio-Resource Bank (AHBB) and the Biobank of Korea University Guro Hospital which are members of Korea Biobank Network supported by the Korean Ministry of Health and Welfare.
Footnotes
Edited by C. Borner
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hyung Chul Lee, Donghee Kang
Supplementary information
The online version of this article (10.1038/s41418-019-0467-6) contains supplementary material, which is available to authorized users.
References
- 1.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28:436–53. doi: 10.1016/j.tcb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Jung SH, Lee HC, Yu DM, Kim BC, Park SM, Lee YS, et al. Heparan sulfation is essential for the prevention of cellular senescence. Cell Death Differ. 2016;23:417–29. doi: 10.1038/cdd.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 5.Krtolica A, Campisi J. Cancer and aging: a model for the cancer promoting effects of the aging stroma. Int J Biochem Cell Biol. 2002;34:1401–14. doi: 10.1016/s1357-2725(02)00053-5. [DOI] [PubMed] [Google Scholar]
- 6.Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–5. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2011;11:S27–31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- 8.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98:12072–77. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D, Hornsby PJ. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 2007;67:3117–26. doi: 10.1158/0008-5472.CAN-06-3452. [DOI] [PubMed] [Google Scholar]
- 10.Coppé JP, Patil CK, Rodier F, Sun Y, Muñoz DP, Godstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–68. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muñoz-Espín D, Cañamero M, Maraver A, Gómez-López G, Contreras J, Murillo-Cuesta S, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155:1104–18. doi: 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–37. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Chongtae, Kang Donghee, Lee Eun Kyung, Lee Jae-Seon. Long Noncoding RNAs and RNA-Binding Proteins in Oxidative Stress, Cellular Senescence, and Age-Related Diseases. Oxidative Medicine and Cellular Longevity. 2017;2017:1–21. doi: 10.1155/2017/2062384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9:e1471. doi: 10.1002/wrna.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, et al. Jarid2 is implicated in the initial xist-induced targeting of PRC2 to the inactive X chromosome. Mol Cell. 2014;53:301–16. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 26.Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelmohsen K, Gorospe M, Noncoding RNA. control of cellular senescence. Wiley Interdiscip Rev RNA. 2015;6:615–29. doi: 10.1002/wrna.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montes M, Lund AH. Emerging roles of lncRNAs in senescence. FEBS J. 2016;283:2414–26. doi: 10.1111/febs.13679. [DOI] [PubMed] [Google Scholar]
- 32.Kent WJ. BLAT—The BLAST-Like alignment tool. Genome Res. 2003;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeCuyer KA, Behien LS, Uhlenbeck OC. Mutants of the bacteriophage MS2 coat protein that alter its coorperative binding to RNA. Biochemistry. 1995;34:10600–06. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- 35.Sun XL, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95:10009–14. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popp MW, Maquat LE. Leveraging rulges of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell. 2016;165:1319–22. doi: 10.1016/j.cell.2016.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–77. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–35. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendell JT, AP Rhys CM, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–22. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- 41.Behm-Ansmant I, Kashima I, Rehwinkel J, Saulière J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–53. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 42.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–27. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–9. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 44.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 45.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. [Google Scholar]
- 46.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–69. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim BC, Lee HC, Lee JJ, Choi CM, Kim DK, Lee JC, et al. Wig1 prevents cellular senescence by regulating p21 mRNA decay through control of RISC recruitment. EMBO J. 2012;31:4289–303. doi: 10.1038/emboj.2012.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HC, Jung SH, Hwang HJ, Kang D, De S, Dudekula DB, et al. WIG1 is crucial for AGO2-mediated ACOT7 mRNA silencing via miRNA-dependent and -independent mechanisms. Nucleic Acids Res. 2017;45:6894–910. doi: 10.1093/nar/gkx307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ju YS, Kim JI, Kim S, Hong D, Park H, Shin JY, et al. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat Genet. 2011;43:745–52. doi: 10.1038/ng.872. [DOI] [PubMed] [Google Scholar]
- 51.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Natl Acad Sci USA. 2012;2:28–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.