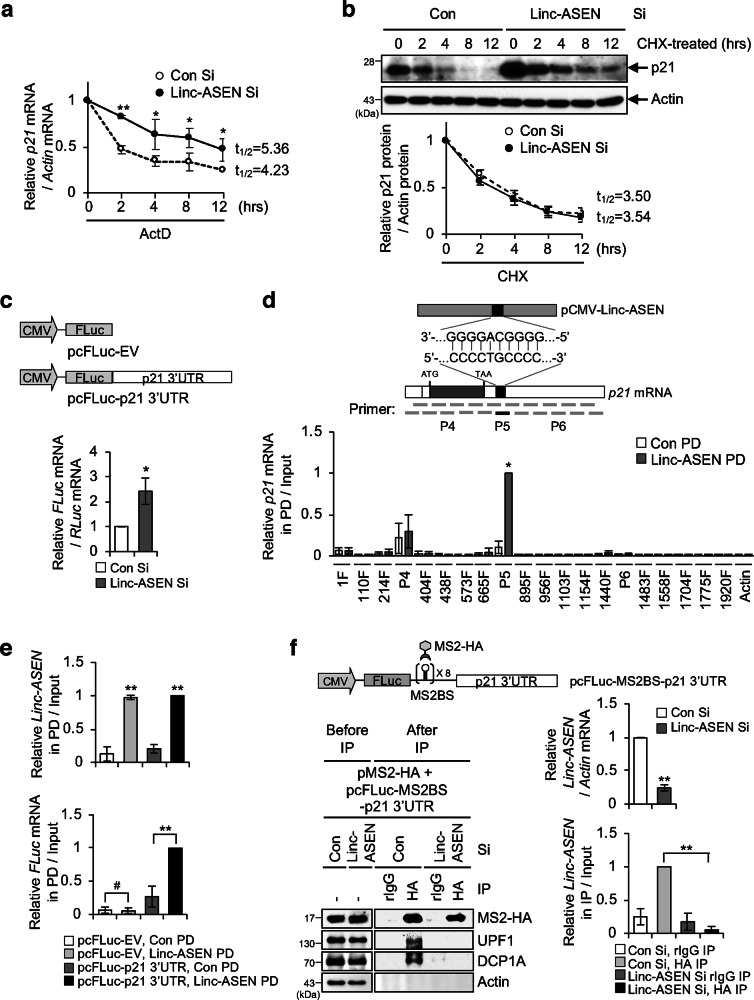
Fig. 5. Linc-ASEN promotes p21 mRNA decay by associating with UPF1 and DCP1A.
a Linc-ASEN silenced MCF7 cells were incubated for the indicated times with 5 mg/ml actinomycin (ActD). RNAs were purified, then subjected to RT-qPCR using p21 mRNA-specific primers. b Linc-ASEN silenced cells were treated with cycloheximide (CHX), after which lysates were prepared at the indicated times for western blot (top). The normalized band intensities were evaluated with ImageJ software (bottom). c Effect of Linc-ASEN depletion on p21 3′UTR reporter mRNA. MCF7 cells were transfected with Linc-ASEN Si prior to transfection with pFL-p21 3′UTR in combination with pRL-CMV as a reference plasmid, and then RT-qPCR analysis was conducted. d Schematic illustration showing primer binding regions of p21 mRNA for ChIRP assay (top). Predicted Linc-ASEN binding site (P5) on p21 mRNA are shown. RNA PD was performed with biotin-Linc-ASEN probes (bottom). p21 mRNA in precipitates was measured by RT-qPCR analysis with 100 nt overlapping interval primers which covers the p21 full-length mRNA. e RNA PD was performed with biotin-Linc-ASEN probes using the FLuc-p21 3′UTR reporter system. Reporter RNA in precipitates was quantified by RT-qPCR analysis using FLuc specific primers. f UPF1 binding on p21 3′UTR is dependent on Linc-ASEN. HEK293T cells were transfected with Linc-ASEN Si prior to transfection with pcFLuc-MS2BS-p21 3'UTR reporter plasmid in combination with pMS2-HA. IP was performed and immunoprecipitates were assessed by western blot and RT-qPCR analyses. In a, c–f, data are represented as means ± SD. *P < 0.05; **P < 0.01; #P > 0.05 (Student’s t-test).