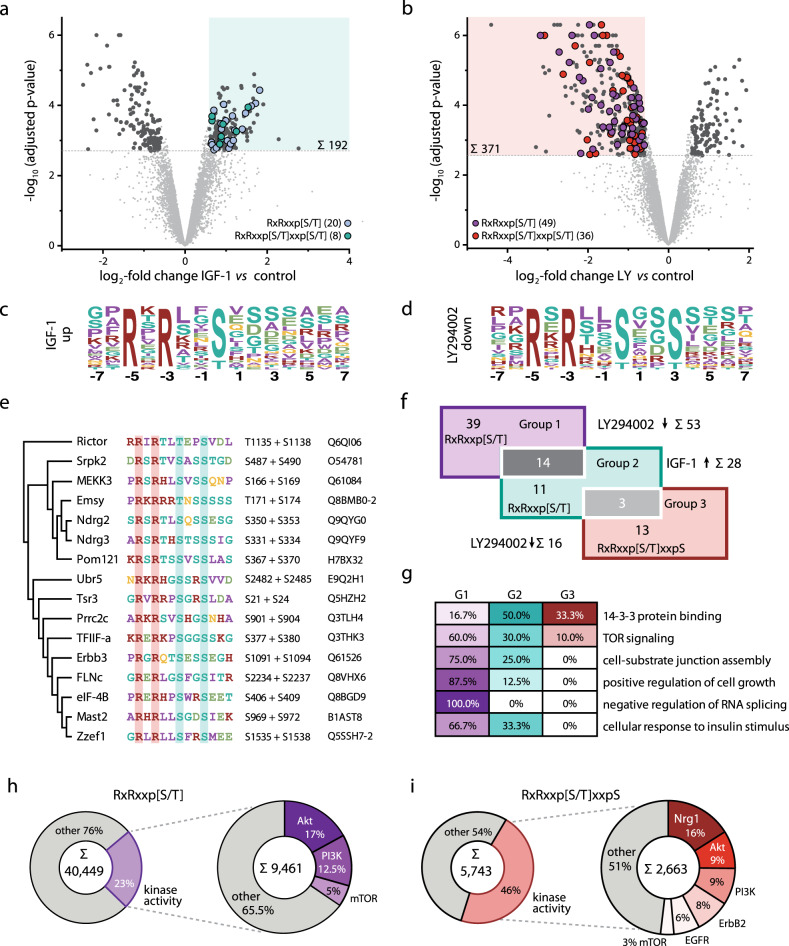
Fig. 2. Identification of an extended basophilic motif by quantitative phosphoproteomics.
a, b Volcano plots of quantified phosphopeptides with localized phosphosites from fully differentiated contracting skeletal myotubes following treatment with IGF-1 (left) or LY294002 (right). Log2-transformed mean SILAC ratios (control/treatment) were plotted against −log10 adjusted p values. Phosphopeptides with a minimum fold change of 1.5 and an adjusted p value lower than 0.05 (n = 3 independent experiments; two-tailed moderated student’s t-test) are shown as dark gray circles. Among these, phosphopeptides are labeled according to their phosphorylation sequence motif as indicated and regions of interest are highlighted in each plot. c, d Motif-X analysis of regulated phosphopeptides. The basophilic motif RxRxxS (c) was 30-fold enriched upon IGF-1 treatment, whereas the extended sequence motif RxRxxSxxS (d) was found to be 123-fold enriched following LY294002 treatment. x, any amino acid. e Clustal Omega analysis of the 16 amino acid sequence window from the proteins comprising regulated peptides with the extended basophilic RxRxxp[S/T]xxpS. f Overlap of unique phosphopeptides comprising the classical basophilic motif RxRxxp[S/T] (Group 1, LY294002 downregulated), RxRxxp[S/T] (Group 2, IGF-1 upregulated), or the extended basophilic motif RxRxxp[S/T]xxpS (Group 3, LY294002 downregulated). g Combined GO enrichment analysis of phosphopeptides of group 1, 2 and 3 (G1–3) shown in (e). Shown are overrepresented pathways with a Benjamini-Hochberg corrected p value ≤ 0.05. h, i Text mining results for interaction partners of proteins comprising the RxRxxp[S/T] (h) or the extended RxRxxp[S/T]xxpS motif (i). Analysis resulted in 40,449 (h) and 5,743 matches (i) of which 9,461 (23%) and 2,663 (46%) were annotated with the term “kinase activity”.