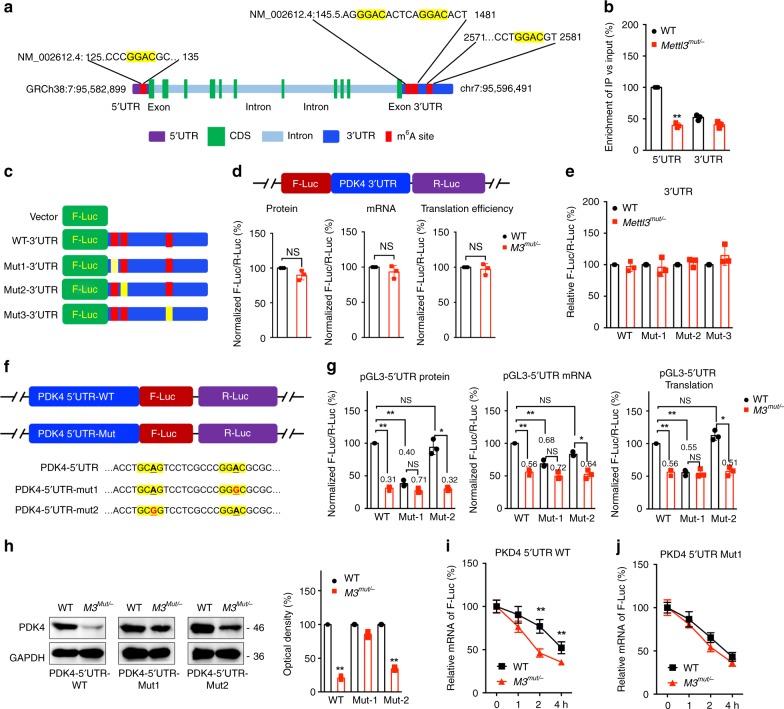
Fig. 4. Methylation sites of PDK4 involved in m6A regulated expression of PDK4.
a Schematic representation of positions of m6A motifs within PDK4 mRNA. b The m6A in 5′UTR or 3′UTR of PDK4 in wild type or Mettl3Mut/- HeLa cells were analyzed by m6A-RIP-qPCR using fragmented RNA. c Schematic representation of mutated (GGAC to GGCC) 3′UTR of pmirGLO vector to investigate the roles of m6A in 3′UTR in PDK4 expression. d Wild-type or Mettl3Mut/- HeLa cells were transfected with pmirGLO-PDK4-3′UTR-WT reporter for 24 h. The protein, mRNA and translation efficiency were determined. e The relative luciferase activity of F-Luc/R-Luc of pmirGLO-3′UTR-WT, or pmirGLO-3′UTR-Mut-1/-2/-3 in wild type and Mettl3Mut/- HeLa cells; f Schematic representation of mutation in 5′UTR to investigate the m6A roles on PDK4 expression. g Wild-type or Mettl3Mut/- HeLa cells were transfected with pGL3-PDK4-5′UTR-WT or pGL3-PDK4-5′UTR-Mut1/2 reporter for 24 h. The protein, mRNA and translation efficiency were determined. h pcDNA-PDK4-5′UTR-WT or pcDNA-PDK4-5′UTR-Mut1/2 was transfected into wild type or Mettl3Mut/- HeLa cells for 24 h. Protein expression was measured by western blot analysis (left) and quantitatively analyzed (right). i, j pcDNA-PDK4-5′UTR-WT (i) or pcDNA-PDK4-5′UTR-Mut1 (j) was transfected into wild type or Mettl3Mut/- HeLa cells for 24 h and then further treated with Act-D for the indicated times. The mRNA of PDK4 was checked by qRT-PCR. Data are presented as the mean ± SD from three independent experiments. *p < 0.05, **p < 0.01, NS, no significant, by two-tailed unpaired Student’s t test for (b) (p < 0.0001), (d) (p > 0.05), and (h) (p < 0.0001 for all), and two-way ANOVA for (c) and (i) (p < 0.001 for all).