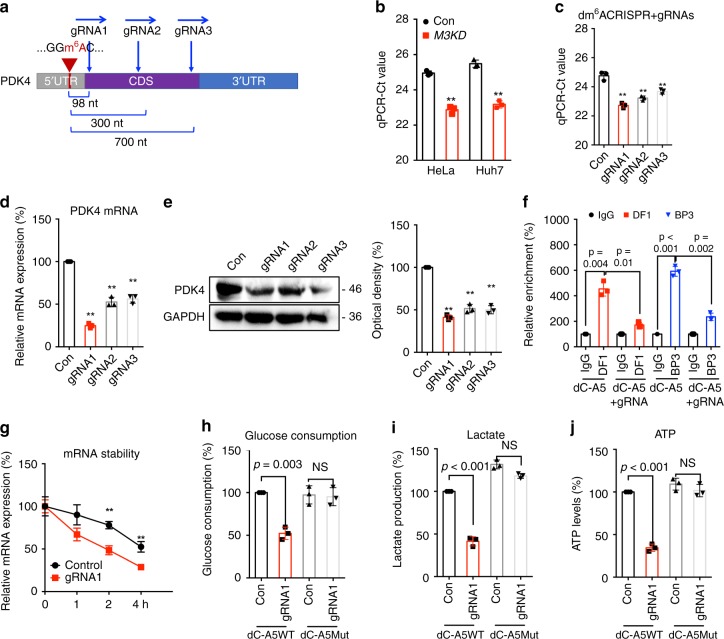
Fig. 6. Targeting m6A of PDK4 by dm6ACRISPR re-programs metabolic of cancer cells.
a Schematic representation of positions of m6A site within PDK4 mRNA and the regions targeted by three gRNAs, respectively. b The threshold cycle (Ct) of qPCR showing SELECT results for detecting m6A site in the potential m6A site of PDK4 5′UTR in Mettl3Mut/- HeLa, sh-Mettl3 Huh7 and their corresponding control cells. c The threshold cycle (Ct) of qPCR showing SELECT results for detecting m6A site in 5′UTR of PDK4 in HeLa cells transfected with dCas13b-ALKBH5 combined with gRNA control or gRNA1/2/3, respectively, for 24 h. d, e The mRNA (d) or protein (e) expression of PDK4 in HeLa cells transfected with dCas13b-ALKBH5 combined with gRNA control or gRNA1/2/3, respectively, for 24 h. f RIP-qPCR analysis of PDK4 mRNA in HeLa cells transfected with dCas13b-ALKBH5 combined with gRNA control (dC-A5) or gRNA for PDK4 (dC-A5 + gRNA) for 24 h by use of antibodies against YTHDF1 and IGF2BP3, respectively. g HeLa cells were transfected with gRNA control, gRNA1 for PDK4, and dCas13b-ALKBH5 for 24 h and then further treated with Act-D for the indicated times. The mRNA of PDK4 was checked by qRT-PCR. h–j The glucose consumption (h), lactate production (i), and ATP levels (j) in HeLa cells transfected with gRNA control, gRNA1 for PDK4, and dCas13b-ALKBH5 or dCas13b-ALKBH5-Mut for 24 h. Data are presented as means ± SD from three independent experiments. **p < 0.01, NS, no significant, by two-tailed unpaired Student’s t test for (b) (p = 0.001 and p = 0.002, respectively), (h–j) one-way ANOVA for (c) (p < 0.001), (d) (p < 0.001), and (e) (p < 0.001), and two-way ANOVA for (f) and (j) (p < 0.001).