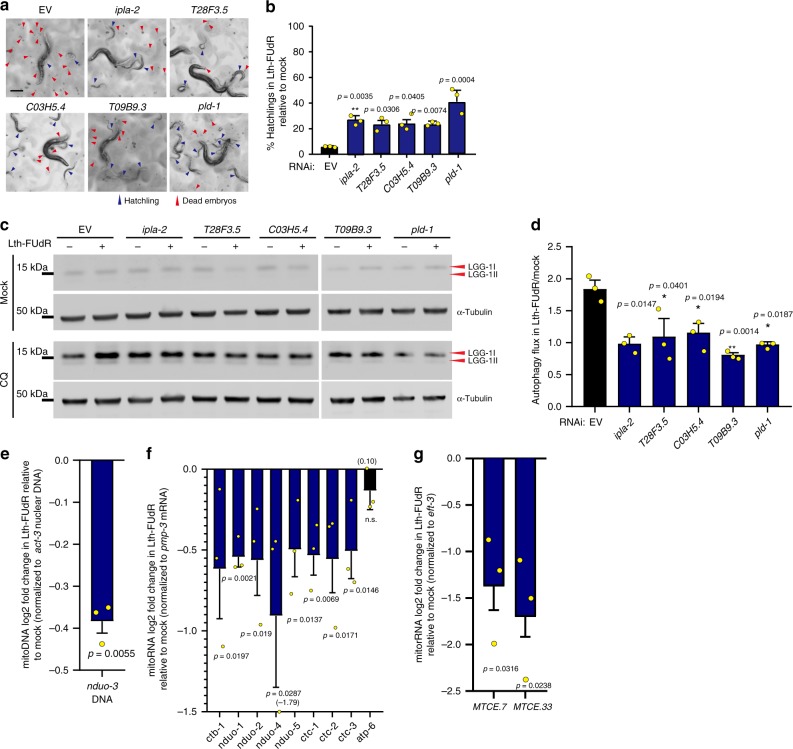
Fig. 6. Autophagy activation in Lth-FUdR depends on mitochondrial lipid metabolism.
Throughout this figure: % hatchlings was analyzed as described in Fig. 1. Statistical significance for %hatchlings quantification was assessed via two-tailed unpaired nonparametric t-test. Statistical significance for western blotting ratio and qPCR fold change was assessed via one-tailed ratio t-test. Data are presented as mean values ± SEM, scale bars = 200 µm. n = # independent biological replicates. Source data are provided as a Source Data file. a Representative images of progeny viability of C. elegans exposed to Lth-FUdR (7.5 μg/mL) while cultured on EORB1 EV or RNAi-targeting mitochondrial genes. n = 3. b Quantification of treatments represented in a. n = 3. c Representative αLGG-1 western blotting analysis of worms exposed to ±Lth-FUdR (7.5 μg/mL) ±8 h of 20 mM chloroquine while cultured on EORB1 EV or RNAi-targeting mitochondrial genes. Data acquisition as described in Methods. d Quantification of autophagy flux of treatments represented in c. Autophagy flux estimation and interpretation as described in Fig. 5h–I, main text, and Methods. n = 3. e qPCR analysis of mitochondrial DNA content (nduo-3) relative to nuclear DNA (act-3) in Lth-FUdR worms relative to mock. n = 3. f qRT-PCR analysis of the expression/stability of mitochondrially encoded mRNAs relative to the nuclearly encoded mRNA pmp-3 in worms treated with Lth-FUdR relative to mock. n = 3. g qRT-PCR analysis of mitochondrially encoded rRNAs normalized to eft-3 (as previously described) in worms treated with Lth-FUdR relative to mock. n = 3.