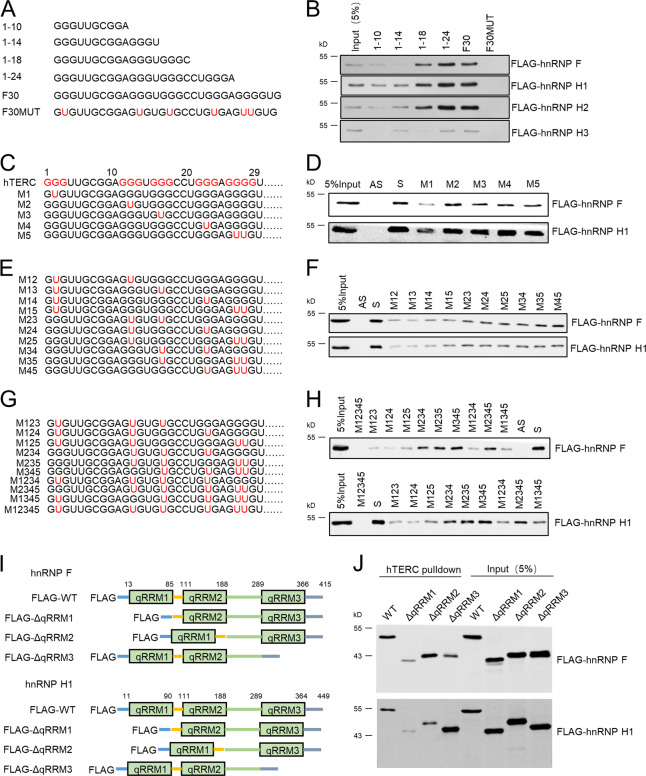
Fig. 2. The guanine runs at 5′-end of hTERC and qRRM1 domain of hnRNP F/H mediate hnRNP F/H-hTERC interaction.
a Nucleotide sequences of each F30 truncated RNAs used for the streptavidin pull-down assay. G to U substitutions made in F30MUT are marked by red. b 3′-biotinylated F30 truncations were incubated with HeLa (over-expressed FLAG-hnRNP F, H1, H2, or H3) whole cell extracts, respectively. The binding of hnRNP F/H to each RNA was assessed by western blotting. c, e and g Nucleotide sequences of each hTERC G to U mutants. G to U substitutions made in 5′-end of hTERC are marked by red. d, f and h Streptavidin pull-down assay was performed, and the binding of hnRNP F/H1 to each RNA was detected by western blot. Biotinylated antisense of hTERC served as a negative control. 5% of whole cell extracts used in reactions as input. i Schematic representation of three qRRM domains of hnRNP F/H1, as well as their truncations lacking of qRRM1, qRRM2, or qRRM3 domain, respectively. j Streptavidin pull-down assays were performed as aforementioned method, and the binding of hTERC to each hnRNP F/H1 truncations was detected by western blot. 5% of whole cell extracts used as input.