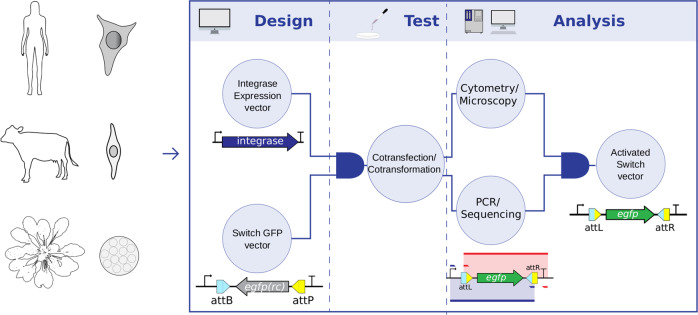
Fig. 1. Strategy overview of the eukaryotic genetic switch system.
The human cell lineage HEK 293T, bovine fibroblasts and A. thaliana protoplasts were the selected model systems. The first step involved the design of two plasmid sets: the integrase expression vectors to express Ints 2, 4, 5, 7, 9, 13, phiC31, or Bxb1 and the switch GFP vectors with the attB and attP recognition sites of the respective Int flanking an egfp coding sequence in a reverse complement (rc) orientation. Acting as a schematic AND gate, combination of the corresponding plasmids of each of the vector sets results in the second step in the process, the test, accomplished by cotransfection or cotransformation assays of mammalian and plant cells, respectively. The third and last step led to the development of analytical methods that include the inputs of an additional schematic AND gate. Microscopy/flow cytometry analyses were used to detect EGFP fluorescence in cells resulting from the flipping action of the integrase. PCR/sequencing was used in the analysis of the egfp coding sequence rotated to the correct forward orientation flanked by the formed attL and attR sites. Both analytical inputs provide evidence of the activated switch vector output. The PCRs used one primer pair to amplify the complete attL site and the entire egfp coding sequence, now in the forward orientation (blue), and a second primer pair to amplify the complete attR site and the egfp sequence (red).