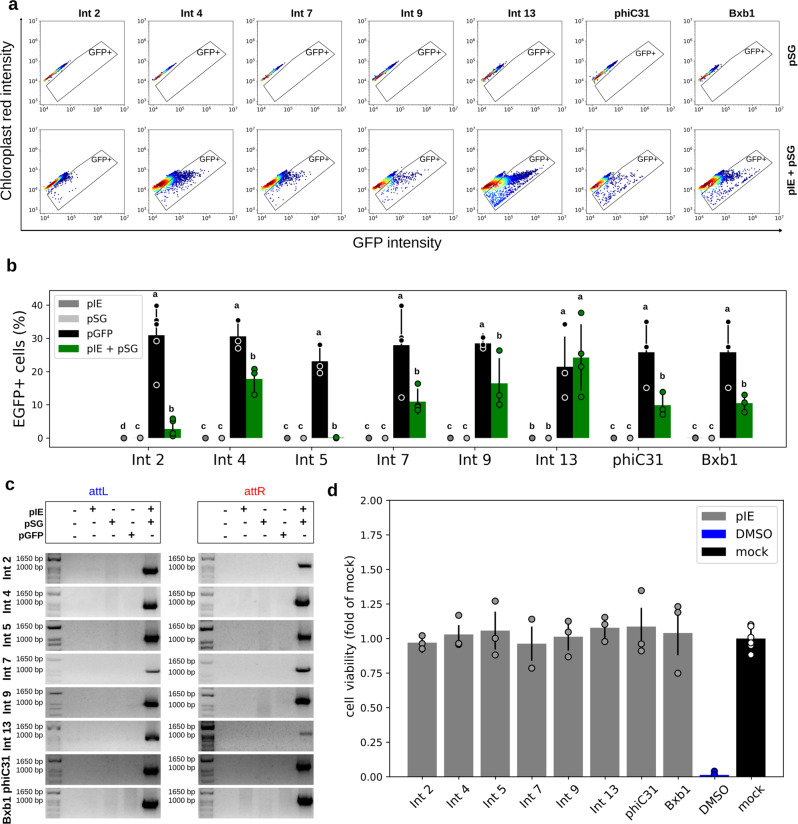
Fig. 4. Functional characterization of the genetic switches in plant protoplasts.
a Flow cytometry distribution of protoplasts at 24 h post cotransformation for Ints 2, 4, 7, 9, 13, phiC31, and Bxb1, the integrases that led to the highest EGFP-expressing cell frequencies. The heat map indicates the scattering of high cell concentrations (warm colors) to low cell concentrations (cool colors). The gate encompasses the EGFP-expressing cell population. b Bar graph plots showing the total average percentage and standard deviation of a cell population expressing EGFP in biological repeat assays (n in Supplementary Table 2) and circles showing the technical triplicate average of each assay on the y axis. The x axis contains the different conditions. For each Int data group, different letters indicate significant differences (p < 0.05). c Amplicons obtained through PCR analysis using two specific primer sets, the first set to verify attL formation and the second set to verify attR (highlighted in Fig. 1). The expected amplicon sizes in the Int test groups varied from 948 to 983 bp for attL and from 1136 to 1132 bp for attR. Negative control cells were transfected with only one of the two vector sets, that is, integrase expression (pIE) or switch GFP (pSG) vectors, plus a mock plasmid. Positive control cells (pGFP) have an egfp sequence in the forward orientation under the control of the CaMV 35S promoter. All the data are representative of three technical and three or more biological replicates. d Bar graph plots showing the viable cells average (circles corresponding to technical replicates averages) and standard deviation normalized with mock plasmid positive cells obtained after FDA assays. DMSO corresponds to the impairment negative control. The cells were transfected with pIE vectors in technical triplicates and three biological replicates (except for Int 7, for which there was three technical and two biological replicates).