Fig. 4. DNA methylation blocks the binding of GATA-1 to the c-Kit CGATA site and attenuates repression in G1E-ER4 cells.
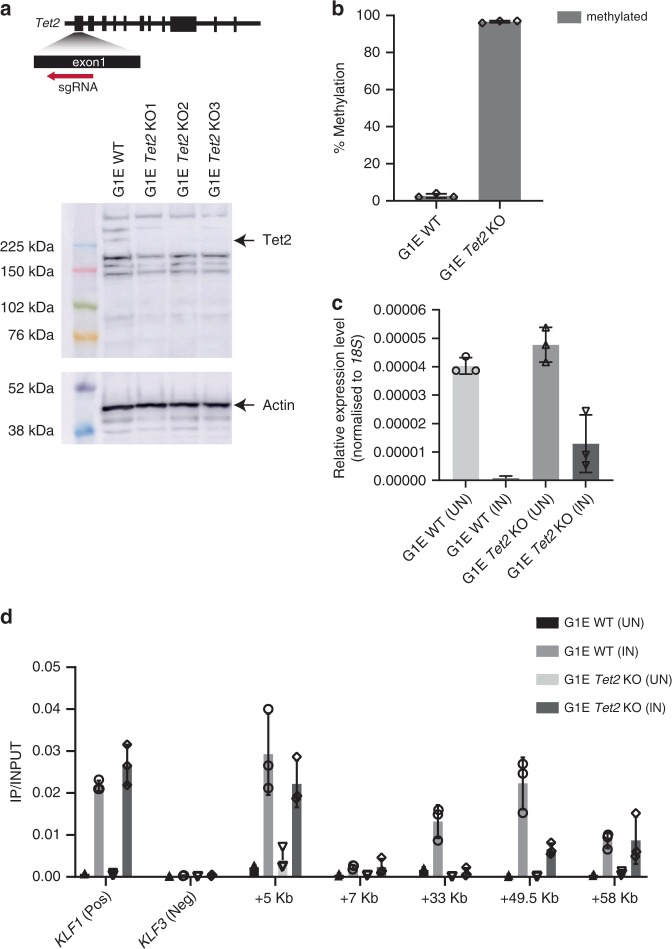
a CRISPR/Cas9 was used to knock out Tet2 in G1E-ER4 cells. Western blot using an antibody to Tet2 identifies clones where Tet2 has been knocked out. A blot using an antibody to actin is shown as a loading control. Three Tet2 knock out clones and clonal wild-type controls were selected for further investigation. The experiment was repeated 3 times independently with similar results. b Methylation level by pyrosequencing of the c-Kit Intron 2 CGATA site in three clonal G1E WT and G1E Tet2 knock out cells (n = 3 biologically independent cells), mean ± standard deviation. c The expression level of c-Kit in G1E WT and G1E Tet2 KO cells by qPCR. Expressed relative to 18S expression, n = 3 biologically independent cells, mean ± standard deviation. Cells were induced with the addition of tamoxifen for 24 h to restore GATA-1-ER fusion proteins in cell nucleus (TAM). Cells incubated with ethanol were used as mock control (NC). d GATA-1 ChIP-qPCR in wild type and G1E Tet2 cells after 24 h of GATA-1 induction by incubating with tamoxifen. KLF1 promoter region was used a positive control (+ve). An upstream region distal to the KLF3 gene was used as a negative control (−ve), the CGATA site at c-Kit intron 2 is the +33 site, n = 3 biologically independent cells, mean ± standard deviation, ChIP-qPCR was repeated two times independently with similar results. Source data are provided as a Source data file.