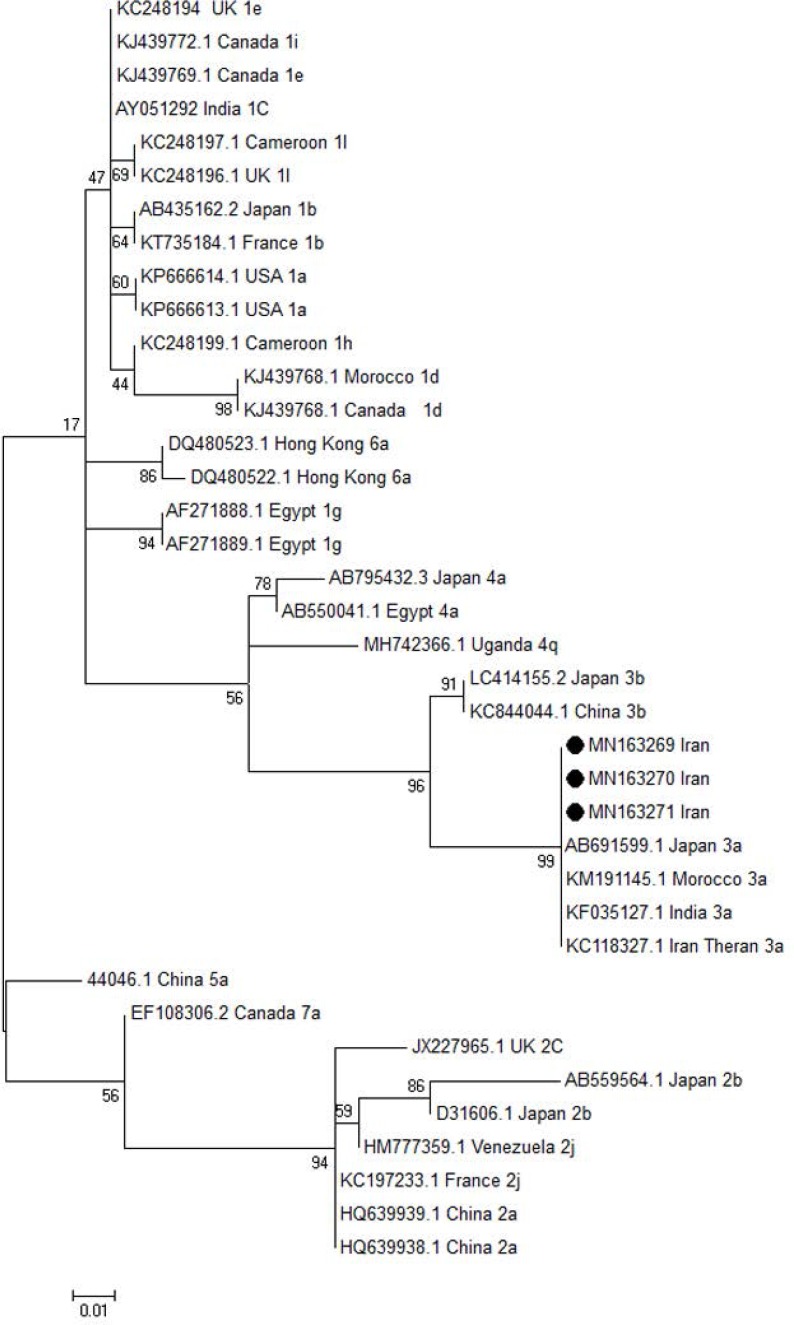
Fig. 1.
The phylogenic tree was constructed for 5UTR region of HCV genome isolated from Ahvaz, Iran. The detected HCV 5UTR region with accession numbers MN163269-71 was compared with different HCV genotypes and subtypes retrieved from GenBank. The maximum likelihood tree method was done under parameters including with the site heterogeneity gamma and invariant sites, phylogenetic distances by Kimura two-parameter model using MEGA 6 software. The Iranian isolated HCV 5UTR region with black circle were clustered with HCV genotype 3a isolated from different regions of the world. The accuracy of the tree was assessed by 1000 bootstrap replicates. The scale bars is equal to 0.01.