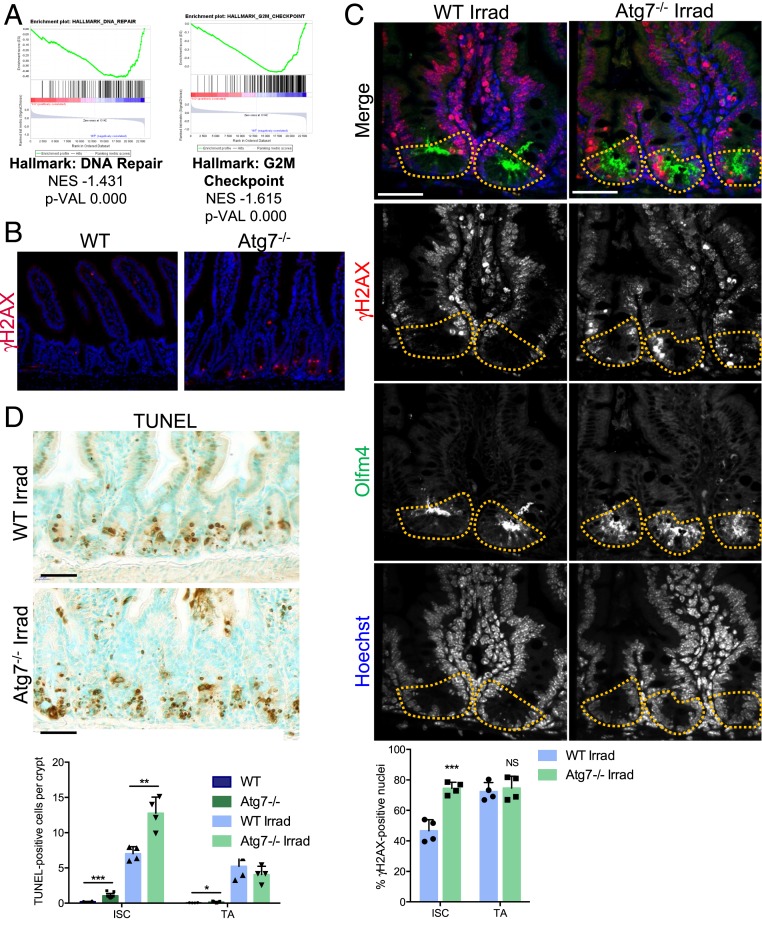
Fig. 3.
Loss of Atg7 disrupts DNA damage repair in Lgr5+ISC. (A) Enrichment plots generated by GSEA for hallmark DNA repair and G2/M checkpoint gene sets based on transcriptomic data for GFPHigh Lgr5+ISC from the crypts of Lgr5Atg7−/− vs. Lgr5WT mice. NES, normalized enrichment score; p-VAL, P value. (B) γH2AX staining showing a lack of epithelial staining in WT crypts and a representative positive zone in the Atg7−/− epithelium. (Scale bars: 50 µm.) (C) Representative z projection (from 20 stacks spanning over 6 µm to include whole nuclei) of combined γH2AX and Olfm4 staining in the crypts of WT and Atg7−/− mice 6 h after 10-Gγ whole-body irradiation. Yellow dashed lines indicate the Lgr5+ISC compartment. The percentage of γH2AX-positive cells was determined on at least 10 randomly selected whole crypts per mouse (n = 4 mice of each genotype). Cells with more than four γH2AX foci in their nuclei were considered γH2AX positive. Olfm4+ cells were considered to be ISC, and the Olfm4− cells above them and below the crypt–villus junction were considered to be TA cells. The data shown are means ± SD. (Scale bars: 50 µm.) (D) Representative TUNEL staining on tissue sections from WT and Atg7−/− mice 6 h after 10-Gγ whole-body irradiation. Determination of the mean number of TUNEL-positive cells per crypt over 50 consecutive whole crypts in 6 control WT mice, 11 control Atg7−/− mice, and 4 irradiated mice of each genotype. The data shown are means ± SD. Significant differences are shown with asterisks. NS, not statistically significant. (Scale bars: 50 µm.) *P < 0.05; **P < 0.01; ***P < 0.005.