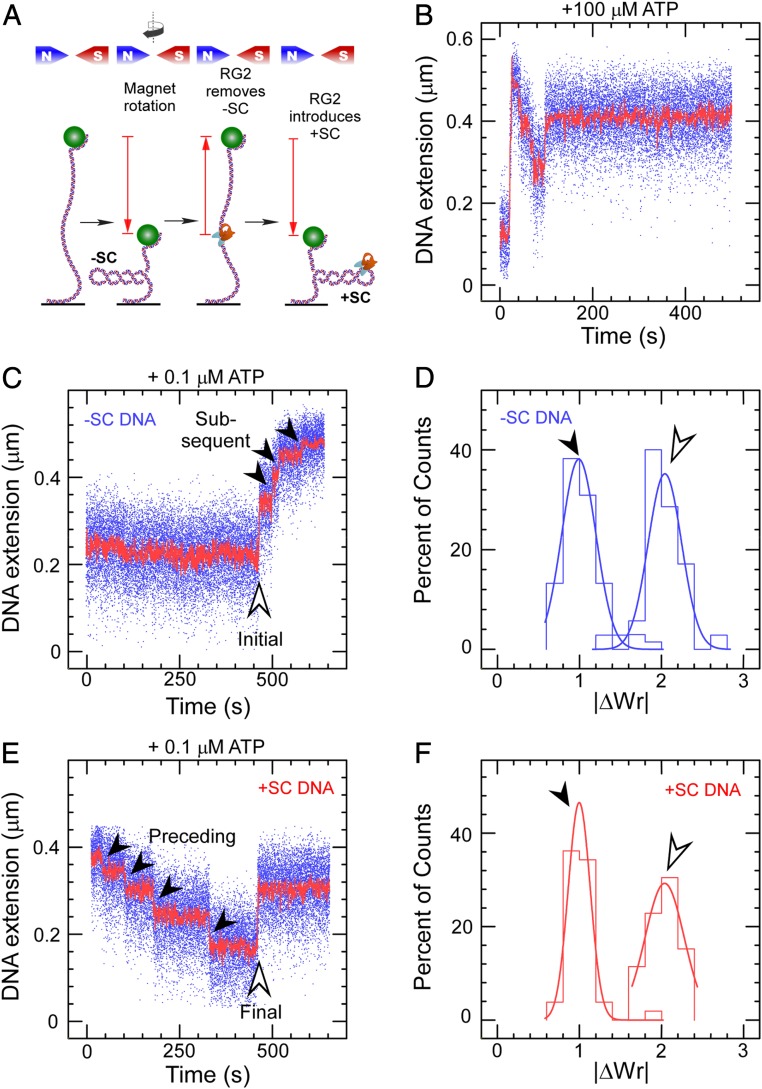
Fig. 1.
Catalytic introduction of positive supercoils by RG2. (A) Sketch of the assay showing DNA tethered between a glass surface and a magnetic microsphere, which can be manipulated with a magnetic trap. Clockwise rotation of the magnets (as seen from above) results in negative DNA supercoils, which reduce DNA extension. RG2 first removes the negative supercoils before introducing net positive supercoiling. (B) Time trace for the extension of a negatively supercoiled DNA exposed to 50 pM RG2 and 100 µM ATP. (C) Time trace for the extension of a negatively supercoiled DNA exposed to RG2 and 0.1 µM ATP. The light arrow highlights the initial interaction, and the filled arrows, the subsequent interactions. (D) Histogram of change in DNA extension observed in C, taking into account both the initial and subsequent interaction observed between RG2 and DNA. Data are fit to Gaussian functions, respectively (solid line), with means ∆Wrinitial = 2.04 ± 0.04 (SEM; n = 35) and ΔWrsubsequent = 0.99 ± 0.03 (SEM; n = 68). E and F are as with the prior two panels but for positively supercoiled DNA, and with means ∆Wrpreceding = 1.00 ± 0.02 (SEM; n = 42) and ∆Wrfinal = −2.03 ± 0.07 (SEM; n = 21).