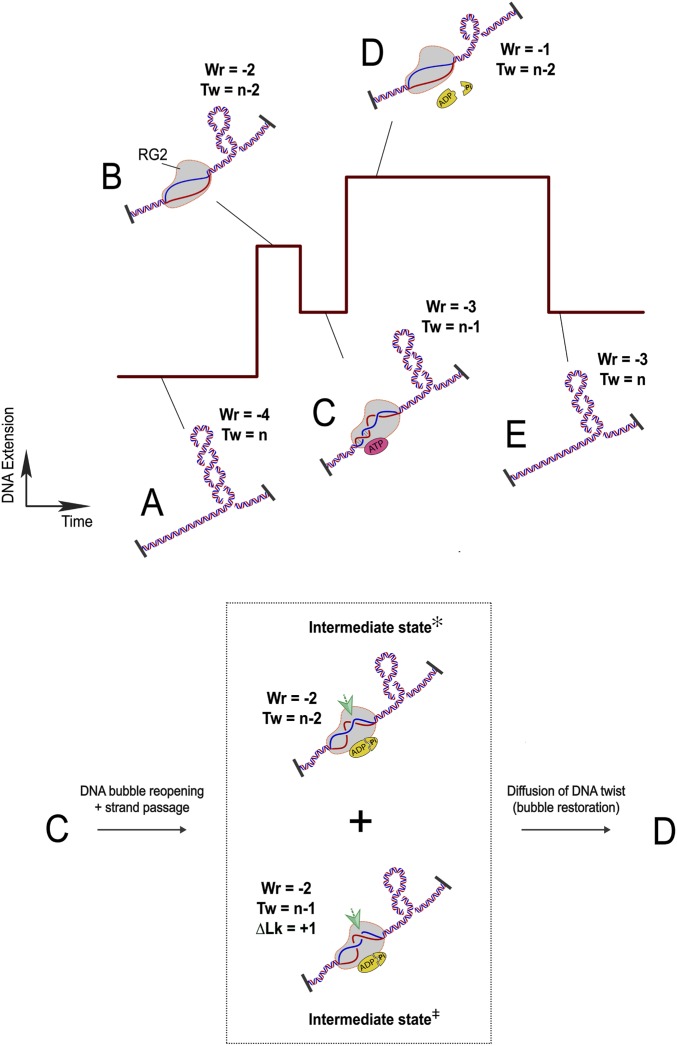
Fig. 5.
Model for the RG cycle. RG2 is viewed as a topological state machine, manipulating DNA twist/writhe in an ordered reaction, through ATP-regulated helicase–topoisomerase coordination. (A) The starting point of an RG2 catalytic cycle is set with a DNA topological domain containing four negative DNA supercoils (Wr = −4; Tw = n). (B) RG2 binding unwinds 20 bp of DNA (Wr = −2; Tw = n − 2). (C) One-half of the 20-base DNA bubble is rewound due to ATP binding of RG2 (Wr = −3; Tw = n − 1). (Inset) In the subsequent ATP hydrolysis and product release stage (C to D), RG2 performs two steps: RG2 conducts a strand-passage reaction at the intersection point indicated by the green arrows (bottom half, Intermediate state⧧; ΔLk = +1 → ΔWr = +1) and RG2 reopens the rewound DNA (top half, Intermediate state✻; ΔTw = −1 → ΔWr = +1). We do not know the order of the two intermediate steps and also the exact conformation of the base pairs. Together these transitions lead to a +2 unit change in DNA writhe. (D) RG2 finally reaches a nucleotide-free state after ATP hydrolysis and product release with the newly produced DNA twist diffused out of the enzyme and thus titrating out one negative supercoil on the DNA. (E) RG2 dissociation from DNA rewinds the 20-base DNA bubble, which returns two negative plectonemes back to the DNA substrate (Tw = −3; Wr = 0). Therefore, a complete ATP cycle by RG2 adds 1 unit of DNA linking number to the DNA (from A–E).