Significance
The speed of the circadian clock is regulated by phosphorylation-regulated degradation of the PER protein. However, this model has recently been challenged by genetic studies in mice and fungi. Here, we provide definitive genetic and biochemical evidence that strongly supports the importance of the phosphoswitch-regulated proteolysis of PER2 in regulating the clock. We generated two independent mouse lines with a point mutation in a casein kinase 1-dependent phosphodegron in PER2. These mice have longer circadian rhythms, increased accumulation of circadian proteins, and perturbed temperature compensation. The findings strongly support the phosphoswitch model of regulated PER2 degradation as a central mechanism controlling the speed of the circadian clock.
Keywords: circadian clock, phosphorylation, knock-in mouse, temperature compensation, Period2
Abstract
Casein kinase 1 (CK1) plays a central role in regulating the period of the circadian clock. In mammals, PER2 protein abundance is regulated by CK1-mediated phosphorylation and proteasomal degradation. On the other hand, recent studies have questioned whether the degradation of the core circadian machinery is a critical step in clock regulation. Prior cell-based studies found that CK1 phosphorylation of PER2 at Ser478 recruits the ubiquitin E3 ligase β-TrCP, leading to PER2 degradation. Creation of this phosphodegron is regulated by a phosphoswitch that is also implicated in temperature compensation. However, in vivo evidence that this phosphodegron influences circadian period is lacking. Here, we generated and analyzed PER2-Ser478Ala knock-in mice. The mice showed longer circadian period in behavioral analysis. Molecularly, mutant PER2 protein accumulated in both the nucleus and cytoplasm of the mouse liver, while Per2 messenger RNA (mRNA) levels were minimally affected. Nuclear PER1, CRY1, and CRY2 proteins also increased, probably due to stabilization of PER2-containing complexes. In mouse embryonic fibroblasts derived from PER2-Ser478Ala::LUC mice, three-phase decay and temperature compensation of the circadian period was perturbed. These data provide direct in vivo evidence for the importance of phosphorylation-regulated PER2 stability in the circadian clock and validate the phosphoswitch in a mouse model.
The cell-autonomous clock systems oscillating with a period of ∼24 h have evolved to coordinate physiological functions with daily environmental changes (1). The molecular mechanism at the heart of the circadian clock is based on transcriptional/translational feedback loops that consist of clock genes and the encoded clock proteins. In the mammalian core clockwork, two proteins, circadian locomotor output cycles kaput (CLOCK) and brain muscle arnt-like 1 (BMAL1), form a heterodimer that binds to the cis-regulatory DNA element E-boxes to activate transcription of Period (Per) and Cryptochrome (Cry) genes. Translated PER and CRY proteins form a multimeric complex with casein kinase 1 (CK1) delta and epsilon (CK1δ/ε) in the cytoplasm and then translocate to the nucleus, where they inhibit the transcriptional activity of CLOCK-BMAL1 complex (2–4). The key molecular components of the clock in all organisms are subject to various posttranslational modifications, most notably phosphorylation and ubiquitination, and disruption of these posttranslational modifications can cause circadian dysfunction (5, 6).
CK1 is an evolutionally conserved core clock component (6). The role of CK1 in the clock is complex. Mutations in CK1, first identified in hamsters (7, 8) and Drosophila (9, 10), can cause both period shortening and lengthening, while pharmacologic inhibition of CK1 consistently causes period lengthening in animals (11–13) and in plants (14). Intriguingly, the kinase activity of CK1 on synthetic peptides is temperature-insensitive (12, 15). This is consistent with a central role for CK1 in temperature compensation of the circadian period, the mechanism that maintains a stable circadian period despite changes in ambient temperature (16–18). In mammals, CK1δ and CK1ε are essential kinases for the circadian clock (19). The first identified mammalian circadian clock mutant, the tau hamster, harbors a semidominant mutation in CK1ε, and tau homozygous animals exhibit markedly shortened locomotor rhythms with a period of 20 h (7, 8). In humans, mutations in CK1δ also cause familial advanced sleep phase (FASP) syndrome and shorten the circadian period (20, 21).
The biochemical function of CK1δ/ε in the clock arises from its stoichiometric tight interaction with and phosphorylation of PER proteins (4, 8, 22, 23). One major consequence of PER2 phosphorylation by CK1δ/ε is the regulation of PER2 stability via the phosphoswitch (11, 17, 18, 24–27). The PER2 phosphoswitch requires at least two phosphorylation domains. Cell-based and biochemical studies have established that Ser478 of PER2 protein is phosphorylated by CK1δ/ε, and Ser478 phosphorylation creates a phosphodegron that recruits an E3 ubiquitin ligase, beta-transducin repeat-containing homolog protein (β-TrCP) (11, 28). β-TrCP is a substrate recognition subunit of the Skp1–Cullin–F-box (SCF) complex that catalyzes formation of a polyubiquitin chain on a substrate targeted for proteasomal degradation. The activity of CK1δ/ε on the S478 phosphodegron is controlled by a second PER2 phosphorylation domain called the FASP region. In this region, CK1δ/ε phosphorylates Ser659, followed by rapid phosphorylation of additional downstream serines in the motif pSxxSxxS. Cell-based assays have demonstrated that this multiphosphorylation of the FASP domain stabilizes PER2 by inhibiting phosphorylation of the Ser478 phosphodegron (17, 18, 24, 26). Indeed, the FASP region is so named because a mutation of human PER2 Ser662 (corresponding to Ser659 in mouse) causes FASP syndrome (25, 29). In the phosphoswitch model, the short-period phenotype common to tau and FASP mutations has been attributed to enhanced phosphorylation of the β-TrCP site, leading to more rapid PER2 degradation. To date, however, there is no in vivo evidence that the β-TrCP site plays a role in determining the circadian period. To test this, we generated two knock-in mouse lines carrying Ser-to-Ala mutation at position 478 of PER2 protein. We find that PER2-Ser478Ala protein is stabilized in the mouse liver, and the mutation caused lengthening of the circadian period of mouse behavioral rhythms. These data provide genetic evidence for the importance of the β-TrCP site in the time-keeping mechanism of the circadian clock in vivo.
Result
The PER2-S478A Mutation Lengthens the Period of Behavioral Rhythms.
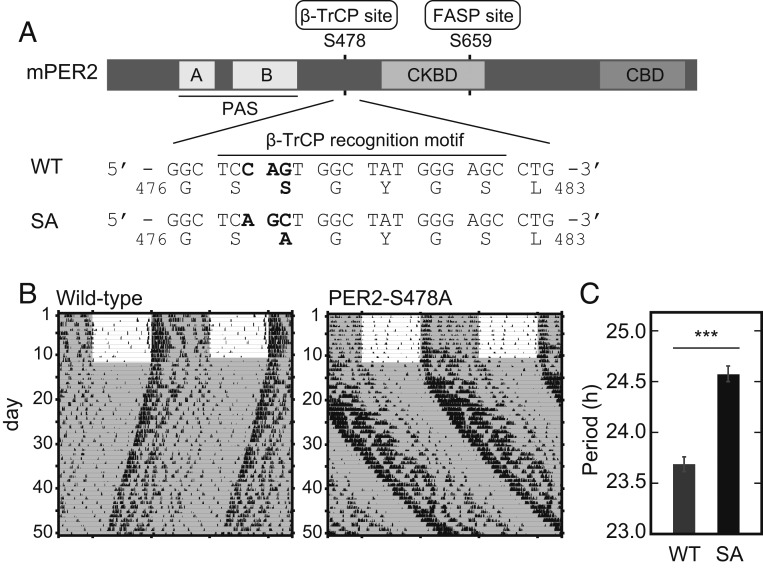
The phosphoswitch model holds that PER2 stability is bidirectionally regulated by two key phosphorylation sites, the β-TrCP site and the FASP site (Fig. 1A). The model also predicts that a mutation of PER2 at the β-TrCP site should result in a more stable protein and a longer circadian period length in vivo. To test this hypothesis, we used CRISPR-Cas9 to generate a mouse with PER2 Ser478Ala (PER2-S478A) (Fig. 1A). PER2-S478A homozygous mice were morphologically normal and fertile and were born in the expected Mendelian ratio. The mutant mice showed robust wheel-running activity rhythms under a 12-h light: 12-h dark (LD) cycle, and no apparent abnormality was observed in the light entrainment (Fig. 1B). On the other hand, when transferred to constant dark (DD) condition, the mutant mice showed ∼1-h longer period of behavioral rhythms (24.58 ± 0.08 h) compared with wild type (WT) (23.67 ± 0.06 h) (Fig. 1C). This is an in vivo demonstration that the Ser478 phosphodegron of PER2 determines the circadian period.
Fig. 1.
The PER2 Ser478Ala mutation lengthens circadian rhythms. (A) The domain structure of PER2 protein is depicted (Upper). The genome sequences in WT and PER2-S478A (SA) knock-in mouse and corresponding amino acid sequences are shown (Lower). CBD, CRY-binding domain; CKBD, CK1-binding domain; PAS, Per-Arnt-Sim domain. (B) Wheel-running activities of representative PER2-S478A homozygous mice and littermate WT mice are shown. Mice were entrained to LD for 2 wk or longer and then exposed to DD. (C) The circadian period of the activity rhythms under the DD condition was determined via a χ2 periodogram procedure based on locomotor activity in days 11 to 24 after the start of DD condition. Mean values ± SEM obtained from 10 WT mice and 12 homozygous PER2-S478A (SA) mice are given. ***P = 4.2 × e−8 (two-sided Student’s t test).
Excessive Accumulation of PER2-S478A Mutant Protein in the Mouse Liver.
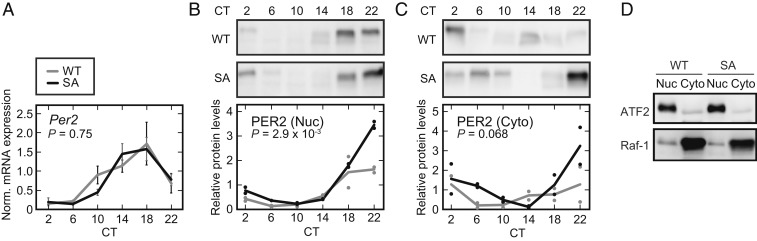
When overexpressed in cultured cells, the PER2-S478A mutant was more stable than the WT PER2 protein (17). Here, we examined the abundance of endogenous Per2 messenger RNA (mRNA) and PER2 protein in the livers of PER2-S478A mice. Livers were harvested at six time points during the second day in the DD condition. Per2 transcripts showed the expected circadian oscillation in both WT and PER2-S478A mice, with no significant difference observed between the genotypes (Fig. 2A). In contrast, peak levels of PER2 protein in both the nucleus and cytoplasm were ∼2-fold higher at circadian time (CT) 22 in PER2-S478A mice (Fig. 2 B and C). Thus, the S478A mutation stabilizes PER2 protein in vivo, similar to what was seen in in vitro overexpression assays (17). It should be noted that, although the peak levels of PER2-S478A protein were higher, PER2-S478A protein was still almost undetectable at CT10 and CT14, suggesting that phosphorylation of Ser478 is required for temporal degradation of PER2 and that another degradation mechanism(s) may be contributing as well (30).
Fig. 2.
PER2 expression levels are posttranscriptionally up-regulated in PER2-S478A mouse liver. (A) Temporal expression of Per2 mRNA normalized by Rps29. Livers were harvested at 4-h intervals followed by real-time RT-qPCR. Mean values ± SEM obtained from three animals of each genotype are given. (B and C) Temporal expression of PER2 proteins in liver. Liver nuclear extracts (B) and cytoplasmic lysates (C) were prepared at 4-h intervals following SDS/PAGE and immunoblotting. Data are represented as dots for individual experiments and as lines for means. P values: two-way ANOVA between WT and PER2-S478A (SA).
The PER2-S478A Mutation Stabilizes CRY Proteins.
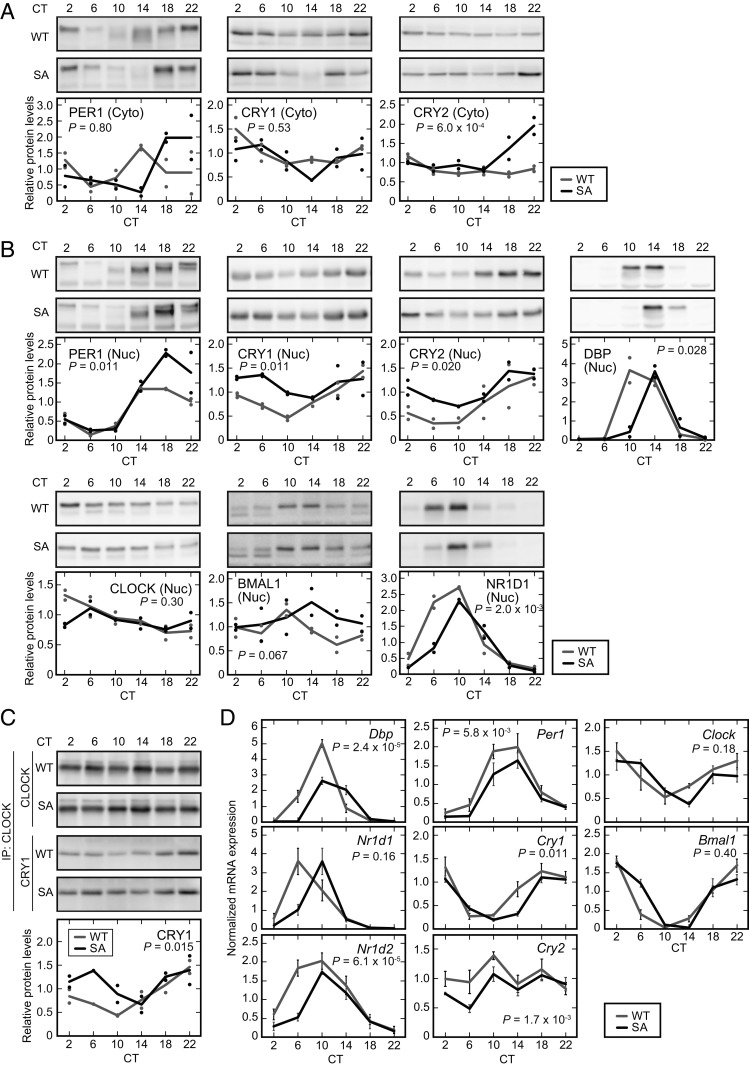
PER2 has been proposed to be rate-limiting for in vivo formation of a stable multiprotein complex containing PER1, CRY1/2, and CK1δ/ε (2, 4) that assembles in the cytoplasm and then translocates to the nucleus. To assess the consequence of stabilization of PER2-S478A protein on these core circadian proteins, we examined the expression of a series of clock proteins in the livers of WT and mutant mice. At the time of peak PER2 abundance (CT22; Fig. 2 B and C), the cytosolic protein levels of PER1 and CRY2 were also increased (Fig. 3A). Lagging by several hours, the nuclear abundance of PER1, CRY1, and CRY2 was also increased in the livers of PER2-S478A mice (Fig. 3B). Thus, stabilized PER2 increases the abundance of the full circadian repressor complex.
Fig. 3.
The PER2-S478A mutation alters the expression profiles of clock genes and proteins in the liver. (A and B) Temporal expression of clock proteins in mouse liver. Liver cytoplasmic lysates (A) and nuclear extracts (B) were prepared at 4-h intervals and analyzed by SDS/PAGE and immunoblotting with indicated antibodies. Data are represented as dots for individual experiments and as lines for means. (C) CRY1 recruitment on the CLOCK complex in the liver nucleus. Liver nuclear extracts were subjected to immunoprecipitation with anti-CLOCK antibody, and the immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. Data are represented as dots for individual experiments and as lines for means. (D) Temporal expression of clock genes. Livers were harvested at 4-h intervals followed by real-time RT-qPCR for the indicated mRNAs. Mean values ± SEM obtained from three animals of each genotype are given. P values: two-way ANOVA between WT and PER2-S478A (SA).
Several studies proposed that PER2 stabilizes CRY proteins by interfering with the interaction between CRYs and FBXL3, an E3 ubiquitin ligase that directs degradation of CRYs (31–33). Up-regulation of CRY protein levels in the PER2-S478A mutant liver (Fig. 3 A and B) suggests that CRYs are stabilized in response to elevation of PER2 protein levels. To test this, we measured the half-lives of CRY1::LUC and CRY2::LUC fusion proteins in HEK 293T cells by monitoring the bioluminescence decay after treatment of cycloheximide (CHX) inhibiting new protein translation. When PER2-S478A was coexpressed, CRY1::LUC and CRY2::LUC were stabilized in a dose-dependent manner and to the same extent as WT PER2 protein (SI Appendix, Fig. S1). Thus, stabilization of PER2 appears to stabilize CRY proteins in both cultured cells and in mouse liver.
PER abundance has been proposed to be the rate-limiting step in the assembly and nuclear entry of the repressor complex (2, 4, 34). However, the nuclear protein levels of CRY1 and CRY2 were still high at CT6 to CT10 (Fig. 3B), when a large proportion of PER2-S478A protein was degraded (Fig. 2B). This suggests that CRY proteins can repress CLOCK-BMAL1 transcriptional activity even in the absence of PER proteins, consistent with a previous study (35). In fact, coimmunoprecipitation analysis showed that CRY1 binds to the CLOCK-containing complex at CT6 to CT10 despite the relative absence of PER2 in the PER2-S478A mutants (Fig. 3C). Furthermore, the mRNA expression profiles of typical E-box–regulated genes, such as albumin D-site binding protein (Dbp), Nr1d1 (also known as Rev-erbA), and Nr1d2 (also known as Rev-erbB), were phase-delayed in the PER2-S478A mutant liver (Fig. 3D). The delayed expression profiles were also observed in RRE-regulated genes Bmal1, Clock, and Cry1 (Fig. 3D). These profiles are consistent with the longer behavioral rhythms of the PER2-S478A mice. Concomitantly, nuclear expression profiles of DBP and NR1D1 proteins were delayed in the mutant mouse liver (Fig. 3B). Overall, our data indicate that the PER2-S478A mutation results in accumulation of circadian repressor complex in the nucleus, leading to delayed derepression of the CLOCK-BMAL1 activator.
The S478A Mutation of PER2 Compromises Temperature Compensation.
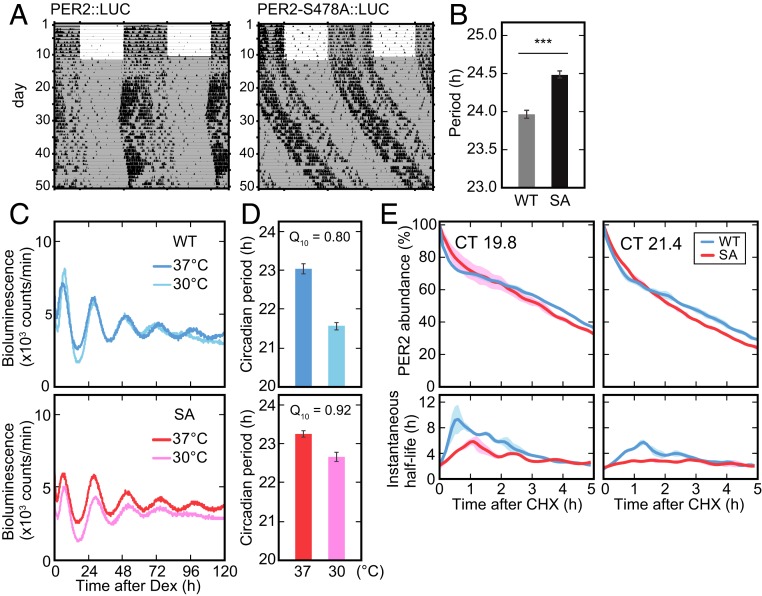
The phosphoswitch model arose from the observation that PER2::LUC degradation occurred in three phases (17): an initial rapid decay (the first phase), a plateau-like slow decay (the second phase), and, finally, a more rapid terminal decay (the third phase). This was visualized by continuous monitoring of bioluminescence signals from PER2::LUC knock-in mouse embryonic fibroblasts (MEFs). To investigate how the S478A mutation affects the three-phase degradation in vivo, we in parallel introduced the S478A mutation into the PER2::LUC allele (PER2-S478A::LUC) encoding a PER2 fusion protein with firefly luciferase (36). In the analysis of the wheel-running activity rhythms, PER2-S478A::LUC mice exhibited a significantly longer circadian period than PER2::LUC control mice (Fig. 4 A and B), similar to that seen in the PER2-S478A mice. We established MEFs from both PER2-S478A::LUC and control PER2::LUC mice.
Fig. 4.
The mutation of PER2 at Ser478 perturbs the phosphoswitch. (A) Wheel-running activities of representative PER2::LUC and PER2-S478A::LUC homozygous mice are shown. Mice were entrained to LD for 2 wk or longer and then exposed to DD. (B) The circadian period of the activity rhythms under the DD condition was determined via a χ2 periodogram procedure based on locomotor activity in days 11 to 24 after the start of DD condition. Mean values ± SEM obtained from 10 PER2::LUC mice (WT) and 10 PER2-S478A::LUC (SA) mice are given. ***P = 5.6 × e−7 (two-sided Student’s t test). (C) Representative recordings of bioluminescence from PER2::LUC or PER2-S478A::LUC MEFs are shown. MEFs were synchronized with dexamethasone, and the bioluminescence was then continually measured in the Lumicycle. (D) The circadian period of cellular rhythms was calculated. Mean values ± SEM obtained from three PER2::LUC MEFs (WT) and three PER2-S478A::LUC (SA) MEFs are given. (E) Bioluminescence from PER2::LUC (WT) and PER2-S478A::LUC (SA) MEFs are shown. MEFs were synchronized with dexamethasone, and the bioluminescence was continually measured in the Lumicycle. CHX (40 µg/mL) was added at CT19.8 (Left) or at CT21.4 (Right). By fitting the exponential curve to the small segment of the decay curves, the instantaneous half-life is calculated (see Materials and Methods for details). Colored ranges represent SEs of curves from their means (n = 3 to 4).
We then tested if disruption of the phosphoswitch by the S478A mutation affects the temperature compensation, which is a key feature of the circadian clock (37). While the rate of most enzymatic reactions doubles or triples with every 10 °C increase in temperature (i.e., temperature coefficient, Q10 = 2 to 3), circadian rhythms are relatively stable in the face of environmental temperature alterations and in fact are often overcompensated, with a Q10 < 1.0. Indeed, in the PER2::LUC MEFs, as the temperature decreased from 37 to 30 °C, the clock sped up ∼1.5 h, giving a Q10 = 0.80 (Fig. 4 C and D and SI Appendix, Fig. S2). However, in the PER2-S478A::LUC MEFs, the clock only sped up ∼0.5 h, for a Q10 of 0.92 (Fig. 4 C and D and SI Appendix, Fig. S2). Thus, temperature compensation was compromised by the S478A mutation, underscoring the importance of this site in the phosphoswitch-regulated temperature compensation.
Finally, we examined whether mutation of the phosphodegron can disrupt the previously observed three-phase decay of PER2::LUC. When protein translation was inhibited by addition of CHX at two distinct time points (CT19.8 and CT21.4) in the PER2 accumulation phase, we again observed three-phase degradation of endogenous PER2::LUC (Fig. 4E). The duration of the second phase was dependent on the time when CHX was added, as previously reported (17). At both time points, the three-phase degradation of PER2 protein was markedly attenuated by the S478A mutation (Fig. 4E), resembling what was seen when CK1 was inhibited (17).
Collectively, these data demonstrate that the phosphodegron in PER2 plays a key role in the phosphoswitch, temperature compensation, and circadian period in vivo.
Discussion
Robust genetic data indicate that CK1δ/ε controls the speed of the circadian clock oscillation, for which multiple potential molecular mechanisms have been proposed. These include nuclear–cytoplasmic shuttling of PER proteins, phosphorylation of PER-associated proteins in the repression complex, phosphorylation of CLOCK to decrease its DNA binding, and a phosphoswitch to regulate PER protein stability (4, 17, 38). We previously demonstrated that phosphorylation of the FASP region inhibited phosphorylation of the β-TrCP phosphodegron, but it has not been experimentally addressed whether the β-TrCP degron actually regulates the circadian period. In the present study, we find that phosphorylation of the β-TrCP site controlling PER2 stability is an essential step in determining the period length in vivo. Mice with the PER2-S478A mutation showed significantly longer behavioral rhythms than control mice (Fig. 1). The period-lengthening effect (by ∼1 h) is similar to those observed in mice injected with a CK1 inhibitor (13) and in transgenic mice carrying human PER2 S662D, a phosphomimic mutant at the FASP site (25). These data show that the β-TrCP site is important for the regulation of behavioral rhythm by the phosphoswitch. Unlike what was observed in hPER2-S662D transgenic mice (25), Per2 mRNA levels were unaltered in the PER2-S478A mutant. The peak levels of PER2 protein were increased in cytosol and nucleus (Fig. 2), consistent with disruption of the Ser478 phosphodegron. Thus, this study provides strong genetic evidence that phosphorylation-regulated degradation of PER2 is indeed a key regulator of the clock speed.
CK1δ and CK1ε are not the only kinases that can regulate PER2 stability. For example, a chemical screening identified CK1α as a regulator of PER protein stability (39). This effect of CK1α may be indirect because CK1α binds to PER proteins with a much lower affinity than CK1δ/ε does (38–40). In Drosophila, CK1α enhances PER degradation elicited by DBT (the CK1δ/ε homolog in Drosophila) (41). CK2 phosphorylates PER2 at Ser53 and promotes PER2 proteasomal degradation in both CK1δ/ε-dependent and -independent pathways (42). PKCα (43) and GRK2 (44) regulate the nuclear entry of PER2 and might affect the stability of PER2. We have previously demonstrated that CK1δ/e phosphorylates Ser478 to control PER2 stability in vitro (11, 17, 45). Here, we showed that mutation of this putative CK1 target site attenuated the degradation of PER2 protein and lengthened the circadian period in vivo. Loss of function of CK1 in Neurospora also lengthened the circadian period (46). The clock genes are not orthologous across species in eukaryotes, but architectures of the molecular clockworks are analogous. FREQUENCY (FRQ), a negative component of the circadian clock in Neurospora, is analogous to PER. CK1 phosphorylates FRQ and decreases its protein stability as well (47). In contrast, recent studies proposed that the binding affinity of CK1 for FRQ, instead of FRQ stability, might be a critical factor for the period determination through phosphorylation of other clock proteins (48, 49). Our data establish a role for the phosphodegron in regulating circadian rhythms but do not rule out an alternative model in which CK1-mediated phosphorylation of other substrates also plays a role for regulation of clock timing.
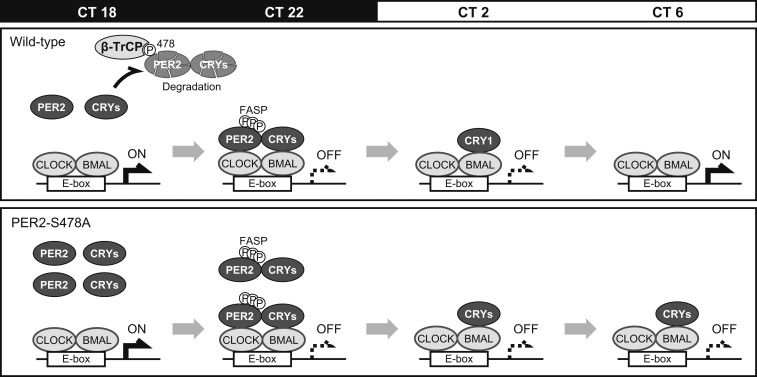
In this study, we propose a model (Fig. 5), in which PER2 Ser478 phosphorylation governs decay of PER/CRY-containing repressor complex that determines restart timing of the E-box–dependent transcriptional activation. The peak level of PER2 protein is regulated by the balance between the competing phosphorylations of stabilizing (FASP) and destabilizing (β-TrCP) sites. The PER2-S478A mutation disrupts this balance and results in excessive PER2 accumulation (Fig. 2) that interferes with the degradation of PER1 and CRY1/2 proteins (Fig. 3). The mature repressive complex enters the nucleus to inhibit CLOCK-BMAL1. The nuclear PER proteins are then degraded, presumably in a β-TrCP site-independent fashion (17). The remaining CRY proteins remain on the CLOCK-BMAL1 activator complex to keep repressing the transcriptional activity in the absence of PERs (35, 50, 51), also contributing to the lengthened period phenotype (Fig. 1).
Fig. 5.
PER2-S478A mutation lengthens the circadian period. PER2 and CRY proteins accumulate during night and disappear before CT6 in WT. In contrast, PER2 and CRY proteins are stabilized and excessively accumulate around CT22 in PER2-S478A mutants. Thereafter, PER2 proteins are degraded by CT2, whereas CRY proteins keep repressing the CLOCK-BMAL complexes.
Temperature compensation (or overcompensation) of the circadian period has been described for more than 60 y (37). Mathematical models predicted that temperature-dependent opposing reactions contribute to the temperature-compensated clockwork (52), although the processes that virtually act in this mechanism have not been understood. We previously demonstrated that the PER2 phosphoswitch may act as the compensating process in mammalian clockwork (17). The phosphoswitch leads to two pools of PER2, one less stable due to degron phosphorylation and the other more stable due to FASP phosphorylation inhibiting degron phosphorylation. Therefore, three-phase degradation can be observed following CHX treatment in PER2::LUC MEFs (17). Here, the three-phase degradation was compromised in PER2-S478A::LUC MEFs, consistent with this model. The mutant MEFs also showed cellular rhythms with a longer period at lower temperature. These results are consistent with the idea that the β-TrCP site is more susceptible to phosphorylation at lower temperature to make the circadian period compensate for temperature changes (17).
Although mutation of the Ser478 phosphodegron increased PER2 stability, PER2 was still degraded between CT22 and CT2 (Fig. 2). Furthermore, the three-phase degradation was indeed perturbed in the PER2-S478A mutant but was not completely abolished (Fig. 4 E, Left). These data suggest an alternative pathway(s) for PER2 degradation. One possible pathway is via an additional β-TrCP site. A potential phosphodegron motif TpSGCSpS is conserved in PER1 (amino acids 121 to 126) and PER2 (amino acids 92 to 97), whereas the primary β-TrCP degron SpSGYGpS (amino acids 477 to 482 in PER2) is not present in PER1 (53). In PER2, the phosphorylation of the primary degron plays a dominant role for β-TrCP–mediated degradation, but a role of the second degron has been reported when the primary degron is mutated (28). In PER2-S478A mutant, this additional degron may therefore contribute to the alternative degradation pathway. Second, it was recently reported that oncoprotein MDM2 targets PER2 for proteasomal degradation, independent of the phosphorylation status of PER2 protein (30). Third, it is possible that β-TrCP recruited to phosphorylated PER1 might contribute to PER2 degradation in the repressor complex. Future studies on detailed mechanisms underlying the PER2 stability control will help to understand the temperature compensation of the circadian clock.
Materials and Methods
Mice.
Mice used in this study (C57BL/6 background) were handled in accordance with the Guidelines for the Care and Use of Laboratory Animals at The University of Tokyo. Mice were housed in cages with free access to commercial chow (CLEA Japan) and tap water. The animals were maintained in a light-tight chamber at a constant temperature (23 ± 1 °C) and humidity (55 ± 10%).
Generation of PER2-S478A Mice and PER2-S478A::LUC Mice.
Four Cas9 guide RNAs targeting the mouse Per2 Ser478 codon were selected for activity testing. Guide RNAs were cloned into a T7 promoter vector, followed by in vitro transcription (HiScribe T7 High Yield RNA Synthesis Kit; New England BioLabs) and spin column purification (RNeasy, Qiagen). Functional testing was performed by incubating Cas9 protein and guide RNA with a PCR-amplified target site, followed by gel electrophoresis to detect target site cleavage. The guide RNA selected for genome editing in embryos was Per2-sg72T (protospacer sequence, 5′-GCGGCTCCAGTGGCTAT-3′). Donor oligonucleotide Per2-S478A-T was used as a donor for point mutation insertion in the Per2 gene (5′-CTTCTACACCTTTCTATGTGCTTTCCCCCCAGCCTGTCCCCCACAGCGGCTCAGCTGGCTATGGGAGCCTGGGCAGTAACGGATCCCACGAACACCTCATGAGCCAGA-3′). SpCas9 protein was expressed and purified by the University of North Carolina (UNC) at Chapel Hill Protein Expression and Purification Core Facility.
Embryos were produced by in vitro fertilization with C57BL/6J females (Jackson Laboratory) as egg donors and Per2luc/luc;CK1ε+/+;CK1δflox/flox males as sperm donors. Single-cell fertilized embryos were microinjected with 400 nM Cas9 protein, 50 ng/μL guide RNA and 50 ng/μL donor oligonucleotide in microinjection buffer: 5 mM tris(hydroxymethyl)aminomethane (Tris)⋅HCl (pH 7.5), 0.1 mM ethylenediaminetetraacetic acid (EDTA). Injected embryos were implanted in recipient pseudopregnant females, and resulting pups were screened by PCR and sequencing for the presence of the Per2S478A allele. Founders harboring the Per2S478A allele were mated to WT C57BL/6J females for germline transmission of the targeted allele and to determine if the targeted allele was in cis with the luciferase knock-in. Note that because the Per2luc allele was produced by gene targeting in embryonic stem cells from strain 129, the Per2 locus in the embryos was heterozygous with 129-derived and C57BL/6J-derived alleles. PCR amplicon sequencing documented polymorphisms in the vicinity of the Ser478 codon that could be used to distinguish the 129 and C57BL/6J alleles. Founders with the S478A mutation in the C57BL/6J allele were used to generate animals with the S478A mutation without the luciferase. Founders with the S478A allele inserted in cis with luciferase in the 129 allele were used to establish the combined S478A-Luc strain.
For the establishment of the Ser478Ala mutant lines, biopsy DNA was genotyped by PCR amplification of a 1,042-base pair region encompassing the Ser478 codon with primers Per2-ScF2 (5′-CGGGTCTCTCTGTGCACTCTTG-3′) and Per2-ScR2 (5′-GATGCTTCCTTCTGTCCTCCA-3′). The PCR product was sequenced with primer Per2-SqR2 (5′-GTGACTTTTGGTTTGAATCTTGC-3′). For genotyping, DNA was extracted from the tail and analyzed by the PCR with the primers Per2-ScF2, Per2-ScFwt (5′-CACAGCGGCTCCAGTGG-3′), Per2-ScRmut (5′-AGGCTCCCATAGCCAGCTG-3′), and Per2-ScR3 (5′-GCTTCTCAGGGAGAGGAACAG-3′).
Behavioral Experiments.
Male mice (6 to 10 wk old) were individually housed in cages equipped with running wheels and were entrained to the 12-h light/12-h dark cycle. After more than 2 wk, wheel revolutions were recorded under the DD condition for 24 d or longer. The locomotor activity was determined by recording the number of wheel revolutions in 5-min bins. The locomotor activities in days 11 to 24 after the start of the DD condition were used for the calculation of the period by a χ2 periodogram procedure with ClockLab software (Actimetrics). All animals recorded in this study showed circadian rhythmicities with P < 0.001.
Cell Culture and Real-Time Monitoring of Rhythmic Gene Expression.
MEFs from PER2::LUC mice and PER2-S478A::LUC mice were maintained at 37 °C under 5% CO2, 95% air in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma) supplemented with 100 units/mL penicillin, 100 µg/mL streptomycin, and 10% fetal bovine serum (FBS). We performed real-time monitoring of luciferase expression as described previously (54) with minor modifications. Briefly, cells were treated with 0.1 µM (final) dexamethasone for 30 min, and then the media were replaced by recording media: phenol red-free DMEM (Sigma) supplemented with 10% FBS, 3.5 g/L glucose, 25 U/mL penicillin, 25 µg/mL streptomycin, 0.1 mM luciferin, and 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes)-NaOH (pH 7.0). The bioluminescence signals were continuously recorded for 5 to 10 d at 37 °C in air with Dish Type Luminescencer, LumiCycle (Actimetrics).
RT-qPCR.
Tissues from 7- to 8-wk-old male mice were collected every 4 h from 38 h after the beginning of the DD condition (projected CT). Livers with three biological replicates were lysed with TRIzol reagent (Invitrogen), and total RNAs were prepared with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. From the total RNAs, complementary DNAs (cDNAs) were synthesized by using Go Script Reverse Transcriptase (Promega) with both anchored (deoxythymine) 15 primer and random oligo primers. For quantification of mRNA levels, the cDNAs were subjected to StepOnePlus Real-Time PCR Systems (Applied Biosystems) by using GoTaq Master Mix (Promega) with the gene specific primers (Table 1).
Table 1.
Primer sequences for RT-qPCR
| Primer | Sequence (5′ to 3′) |
| mRps29-Fw | TGAAGGCAAGATGGGTCAC |
| mRps29-Rv | GCACATGTTCAGCCCGTATT |
| mPer1-Fw | CAGGCTAACCAGGAATATTACCAGC |
| mPer1-Rv | CACAGCCACAGAGAAGGTGTCCTGG |
| mPer2-Fw | GGCTTCACCATGCCTGTTGT |
| mPer2-Rv | GGAGTTATTTCGGAGGCAAGTGT |
| mCry1-Fw | CCCAGGCTTTTCAAGGAATGGAACA |
| mCry1-Rv | TCTCATCATGGTCATCAGACAGAGG |
| mCry2-Fw | GGGACTCTGTCTATTGGCATCTG |
| mCry2-Rv | GTCACTCTAGCCCGCTTGGT |
| mClock-Fw | CCTATCCTACCTTGGCCACACA |
| mClock-Rv | TCCCGTGGAGCAACCTAGAT |
| mArntl-Fw | GCAGTGCCACTGACTACCAAGA |
| mArntl-Rv | TCCTGGACATTGCATTGCAT |
| mDbp-Fw | AATGACCTTTGAACCTGATCCCGCT |
| mDbp-Rv | GCTCCAGTACTTCTCATCCTTCTGT |
| mNr1d1-Fw | CGTTCGCATCAATCGCAACC |
| mNr1d1-Rv | GATGTGGAGTAGGTGAGGTC |
| mNr1d2-Fw | ACGGATTCCCAGGAACATGG |
| mNr1d2-Rv | CCTCCAGTGTTGCACAGGTA |
Fw, forward; Rv, reverse.
Preparation of Nuclear and Cytosolic Fractions of Mice Liver.
The nuclear proteins and cytosolic proteins were isolated as previously described (55). Briefly, the mouse tissue (1 g wet weight) was washed with ice-cold phosphate-buffered saline and homogenized on ice with 11 mL of ice-cold buffer A (10 mM Hepes-NaOH [pH 7.8], 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 4 µg/mL aprotinin, 4 µg/mL leupeptin, 50 mM NaF, and 1 mM Na3VO4). The homogenate was centrifuged (5 min each, 700 × g), and the supernatant was used as “cytosolic fraction.” The resultant precipitate was rinsed and centrifuged (5 min each, 700 × g). The precipitate was resuspended in 2 mL of ice-cold buffer C (20 mM Hepes-NaOH [pH 7.8], 400 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 2% [vol/vol] glycerol, 1 mM DTT, 1 mM PMSF, 4 µg/mL aprotinin, 4 µg/mL leupeptin, 50 mM NaF, and 1 mM Na3VO4). After being gently mixed at 4 °C for 1 h, the suspension was centrifuged (30 min each, 21,600 × g) twice, and the final supernatant was used as “nuclear fraction.”
Immunoprecipitation.
Immunoprecipitation was performed as previously described (55) with some modifications. Nuclear fraction of mouse liver cells was diluted with 2 volumes of buffer D1 (20 mM Hepes-NaOH [pH 7.8], 5.5 mM NaCl, 1 mM EDTA, 6.5% [vol/vol] glycerol, 1.5% [vol/vol] Triton X-100, 1 mM DTT, 1 mM PMSF, 4 µg/mL aprotinin, 4 µg/mL leupeptin, 50 mM NaF, and 1 mM Na3VO4). The lysate was incubated at 4 °C for 30 min with anti-CLOCK antibody (55) (CLNT1; D334-3; Medical & Biochemical Laboratories). Protein G-Sepharose 4 Fast Flow (Amersham Biosciences) was then added to this mixture, and the resultant was mixed gently 4 °C for 1 h. The beads were collected by centrifugation for 5 min at 2,400 × g as the immunoprecipitates.
Antibodies and Immunoblot Analysis.
Liver nuclear and cytosolic proteins separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS/PAGE) were transferred to polyvinylidene difluoride membrane (Millipore). The blots were blocked in a blocking solution—1% (wt/vol) skim milk in TBS (50 mM Tris⋅HCl, 140 mM NaCl, 1 mM MgCl2 [pH 7.4])—for 1 h at 37 °C and then incubated overnight at 4 °C with a primary antibody in the blocking solution. The signals were visualized by an enhanced chemiluminescence detection system (PerkinElmer Life Sciences). The blot membrane was subjected to densitometric scanning, and the band intensities were quantified using ImageQuant TL (GE Healthcare). The primary antibodies used in this study were as follows: anti-CLOCK (55) (CLSP3; D333-3; Medical & Biological Laboratories), anti-ARNTL (55) (B1BH2; D335-3; Medical & Biological Laboratories), anti-PER1 (PM091; Medical & Biological Laboratories), anti-PER2 (PM083; Medical & Biological Laboratories), anti-CRY1 (PM081; Medical & Biological Laboratories), anti-CRY2 (PM082; Medical & Biological Laboratories), anti-DBP (PM079; Medical & Biological Laboratories), anti-NR1D1 (PM092; Medical & Biological Laboratories), anti–Raf-1 (C-12; sc-133; Santa Cruz Biotechnology), and anti-ATF2 (C-19; sc-187; Santa Cruz Biotechnology).
Degradation Assay.
Degradation assays were performed as previously described (56) with minor modifications. Plasmids driving expression of CRY1::LUC or CRY2::LUC were cotransfected with myc-PER2 or myc-PER2-S478A plasmids in HEK 293T cells. After 48 h, the transfected cells were treated with 100 µg/mL CHX (Nakalai tesque) before monitoring the bioluminescence in LumiCEC (Churitsu) or were harvested for immunoblotting. Degradation assay in cultured MEFs was performed in essentially the same way except that DNA was not transfected.
To calculate half-lives of CRY1::LUC and CRY2::LUC, each set of bioluminescence data was fitted by two-term exponential model:
where x represents time, and ai, b, and τi are constant values such as initial quantities, baseline, and half-lives, respectively. These values were computed by linear least-square fittings. The fact that the data were fitted well by two-term exponential model indicates CRY1::LUC and CRY2::LUC can exist at least in two states with different stability. The half-life of unstable form (τ1) was likely to be independent of PER2 abundance, whereas that of stable form (τ2) was significantly increased in a PER2-dependent manner. The accumulation of CRY proteins in PER2-S478A mice liver can be explained by stabilization of the stable form of CRY proteins; thus, we described τ2 as the half-lives in SI Appendix, Fig. S1.
To calculate the instantaneous half-life of PER2, we divide the PER2 decay curve into segments of 1 h with a sliding window, which is moved at increments of 0.1 h, as done by Zhou et al. (17). Then, each segment of the decay curve is fitted to the exponential decay curve (). This allows us to track the change of instantaneous half-life of PER2 (), which is described in Fig. 4 E, Bottom.
Data Availability.
All data generated or analyzed in this study are included in the main text and SI Appendix.
Supplementary Material
Acknowledgments
We thank Dale Cowley of the TransViragen/UNC Chapel Hill Animal Models Core for design and production of the Ser478Ala mutant mice and Dr. Atsu Aiba at the University of Tokyo for help with mouse embryo freezing and the embryo transfer. This work was partially supported by the Singapore’s National Medical Research Council/Clinician-Scientist Individual Research Grant (NMRC/CIRG/1465/2017) (to D.M.V.), by Grants-in-Aid for Specially Promoted Research (17H06096) (to Y.F.) and for Scientific Research (B) (25440041) (to H.Y.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, by the Precursory Research for Innovative Medical Care (PRIME) from Japan Agency for Medical Research and Development (17937210) (to H.Y.), and by the Human Frontiers Science Program Organization (RGY0063/2017) (to J. K. K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000266117/-/DCSupplemental.
References
- 1.Reppert S. M., Weaver D. R., Coordination of circadian timing in mammals. Nature 418, 935–941 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Lee C., Etchegaray J. P., Cagampang F. R., Loudon A. S., Reppert S. M., Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aryal R. P., et al. , Macromolecular assemblies of the mammalian circadian clock. Mol. Cell 67, 770–782.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirano A., Fu Y. H., Ptáček L. J., The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 23, 1053–1060 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Gallego M., Virshup D. M., Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8, 139–148 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Ralph M., Menaker M., A mutation of the circadian system in golden hamsters. Science 241, 1225–1227 (1988). [DOI] [PubMed] [Google Scholar]
- 8.Lowrey P. L., et al. , Positional syntenic cloning and functional characterization of the mammalian circadian mutation. Science 288, 483–491 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloss B., et al. , The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94, 97–107 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Price J. L., et al. , double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Eide E. J., et al. , Control of mammalian circadian rhythm by CKI ε -regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 25, 2795–2807 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isojima Y., et al. , CKIepsilon/δ-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 106, 15744–15749 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng Q.-J., et al. , Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc. Natl. Acad. Sci. U.S.A. 107, 15240–15245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uehara T. N., et al. , Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 116, 11528–11536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shinohara Y., et al. , Temperature-sensitive substrate and product binding underlie temperature-compensated phosphorylation in the clock. Mol. Cell 67, 783–798.e20 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Tosini G., Menaker M., The tau mutation affects temperature compensation of hamster retinal circadian oscillators. Neuroreport 9, 1001–1005 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Zhou M., Kim J. K., Eng G. W., Forger D. B., Virshup D. M., A Period2 phosphoswitch regulates and temperature compensates circadian period. Mol. Cell 60, 77–88 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Narasimamurthy R., et al. , CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. Proc. Natl. Acad. Sci. U.S.A. 115, 5986–5991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H., Chen R., Lee Y., Yoo S., Lee C., Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 106, 21359–21364 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y., et al. , Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434, 640–644 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Brennan K. C., et al. , Casein kinase iδ mutations in familial migraine and advanced sleep phase. Sci. Transl. Med. 5, 183ra56 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keesler G. A., et al. , Phosphorylation and destabilization of human period I clock protein by human casein kinase I ε. Neuroreport 11, 951–955 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Maywood E. S., Chesham J. E., Smyllie N. J., Hastings M. H., The Tau mutation of casein kinase 1ε sets the period of the mammalian pacemaker via regulation of Period1 or Period2 clock proteins. J. Biol. Rhythms 29, 110–118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanselow K., et al. , Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 20, 2660–2672 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y., et al. , Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell 128, 59–70 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shanware N. P., et al. , Casein kinase 1-dependent phosphorylation of familial advanced sleep phase syndrome-associated residues controls PERIOD 2 stability. J. Biol. Chem. 286, 12766–12774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fustin J. M. et al., Two Ck1δ transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 115, 5980–5985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohsaki K., et al. , The role of β-TrCP1 and β-TrCP2 in circadian rhythm generation by mediating degradation of clock protein PER2. J. Biochem. 144, 609–618 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Toh K. L., et al. , An hPer2 phosphorylation site mutation in familiar advanced sleep phase syndrome. Science 291, 1040–1043 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Liu J., et al. , Distinct control of PERIOD2 degradation and circadian rhythms by the oncoprotein and ubiquitin ligase MDM2. Sci. Signal 11, eaau0715 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Chen R., et al. , Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol. Cell 36, 417–430 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nangle S. N., et al. , Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. eLife 3, e03674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xing W., et al. , SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye R., et al. , Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev. 28, 1989–1998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koike N., et al. , Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo S. H., et al. , PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hastings J. W., Sweeney B. M., On the mechanism of temperature independence in a biological clock. Proc. Natl. Acad. Sci. U.S.A. 43, 804–811 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vielhaber E., Eide E., Rivers A., Gao Z. H., Virshup D. M., Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol. Cell. Biol. 20, 4888–4899 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirota T., et al. , High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol. 8, e1000559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dahlberg C. L., Nguyen E. Z., Goodlett D., Kimelman D., Interactions between Casein kinase iepsilon (CKIepsilon) and two substrates from disparate signaling pathways reveal mechanisms for substrate-kinase specificity. PLoS One 4, e4766 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam V. H., et al. , CK1α collaborates with DOUBLETIME to regulate PERIOD function in the Drosophila circadian clock. J. Neurosci. 38, 10631–10643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuchiya Y., et al. , Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci. Signal. 2, ra26 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Jakubcakova V., et al. , Light entrainment of the mammalian circadian clock by a PRKCA-dependent posttranslational mechanism. Neuron 54, 831–843 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Mehta N., et al. , GRK2 fine-tunes circadian clock speed and entrainment via transcriptional and post-translational control of PERIOD proteins. Cell Rep. 12, 1272–1288 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Philpott J. M., et al. , Casein kinase 1 dynamics underlie substrate selectivity and the PER2 circadian phosphoswitch. eLife 9, 1–24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He Q., et al. , CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 20, 2552–2565 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Querfurth C., et al. , Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol. Cell 43, 713–722 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Larrondo L. F., et al. , Decoupling circadian clock protein turnover from circadian period determination. Science 347, 1257277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X., et al. , FRQ-CK1 interaction determines the period of circadian rhythms in Neurospora. Nat. Commun. 10, 4352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shearman L. P., et al. , Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Ye R., Selby C. P., Ozturk N., Annayev Y., Sancar A., Biochemical analysis of the canonical model for the mammalian circadian clock. J. Biol. Chem. 286, 25891–25902 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruoff P., Introducing temperature‐compensation in any reaction kinetic oscillator model. J Interdisiplinary Cycle Res 23, 92–99 (1992). [Google Scholar]
- 53.Shirogane T., Jin J., Ang X. L., Harper J. W., SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 280, 26863–26872 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Imamura K., et al. , ASK family kinases mediate cellular stress and redox signaling to circadian clock. Proc. Natl. Acad. Sci. U.S.A. 115, 3646–3651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshitane H., et al. , Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol. Cell. Biol. 29, 3675–3686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirano A., et al. , FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152, 1106–1118 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in this study are included in the main text and SI Appendix.