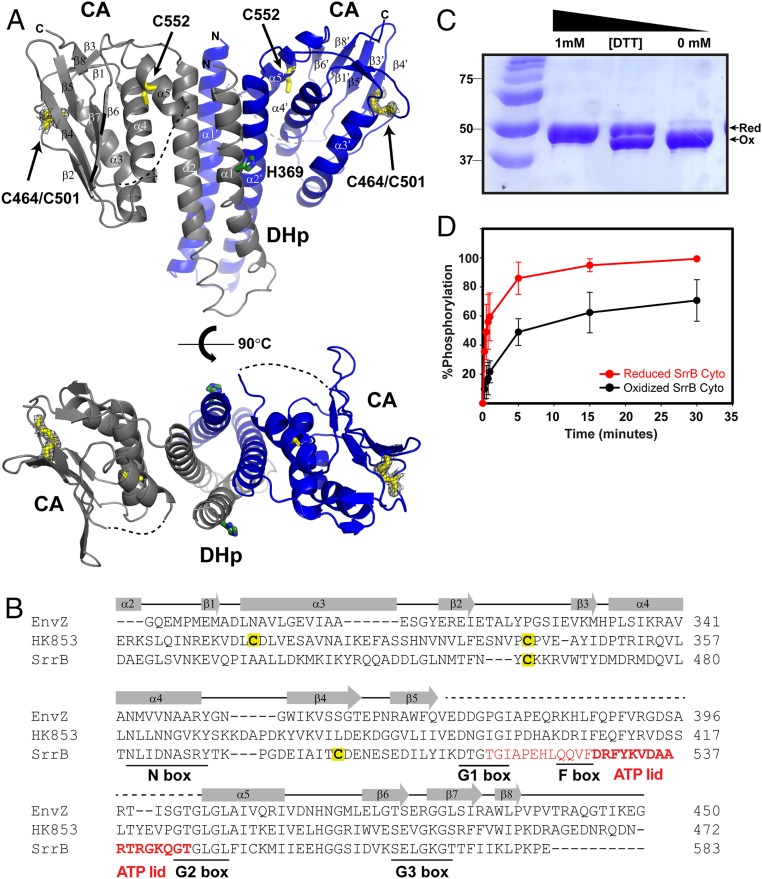
Fig. 2.
The SrrB histidine kinase activity is sensitive to the cysteine redox state. (A) The crystal structure of oxidized (-DTT) SrrB catalytic DHp-CA region refined to 2.0 Å (PDB ID code 6PAJ). The phosphorylated histidine residue (H369) is colored in green, and the cysteine residues are in yellow. The missing electron densities for residues 517 to 544 are shown as a dashed line. (Bottom) Top-down view of the DHp-CA region. Each monomer is shown in gray and blue, respectively. The 2Fo-Fc electron density map of the disulfide bond between C464 and C501 is shown in gray wire at contour level 1σ. (B) A structure-based sequence alignment of E. coli EnvZ (PDB ID code 4KP4; NCBI accession no. 584579776), T. maritima HK853 (PDB ID code 3DGE; NCBI accession no. 251836869), and S. aureus SrrB (NCBI accession no. 123003464) is shown with conserved N, G1, F, G2, and G3 box residues underlined. The residues 517 to 544, which are missing in the SrrB crystal structure, are colored in red, and residues forming the ATP-lid are in bold. (C) SDS/PAGE gel of SrrB Cyto showing redox regulation of the intramolecular disulfide bond by DTT. DTT (0 to 1 mM) was added to the reaction buffer containing SrrB Cyto, run on 12% SDS/PAGE gel, and stained with Coomassie blue. (D) Redox-dependent autophosphorylation of SrrB Cyto. Phosphorylated SrrB Cyto was quantified by using ImageJ and plotted in SigmaPlot. The autophosphorylation reactions of reduced and oxidized SrrB Cyto were done in triplicate.