Figure 3.
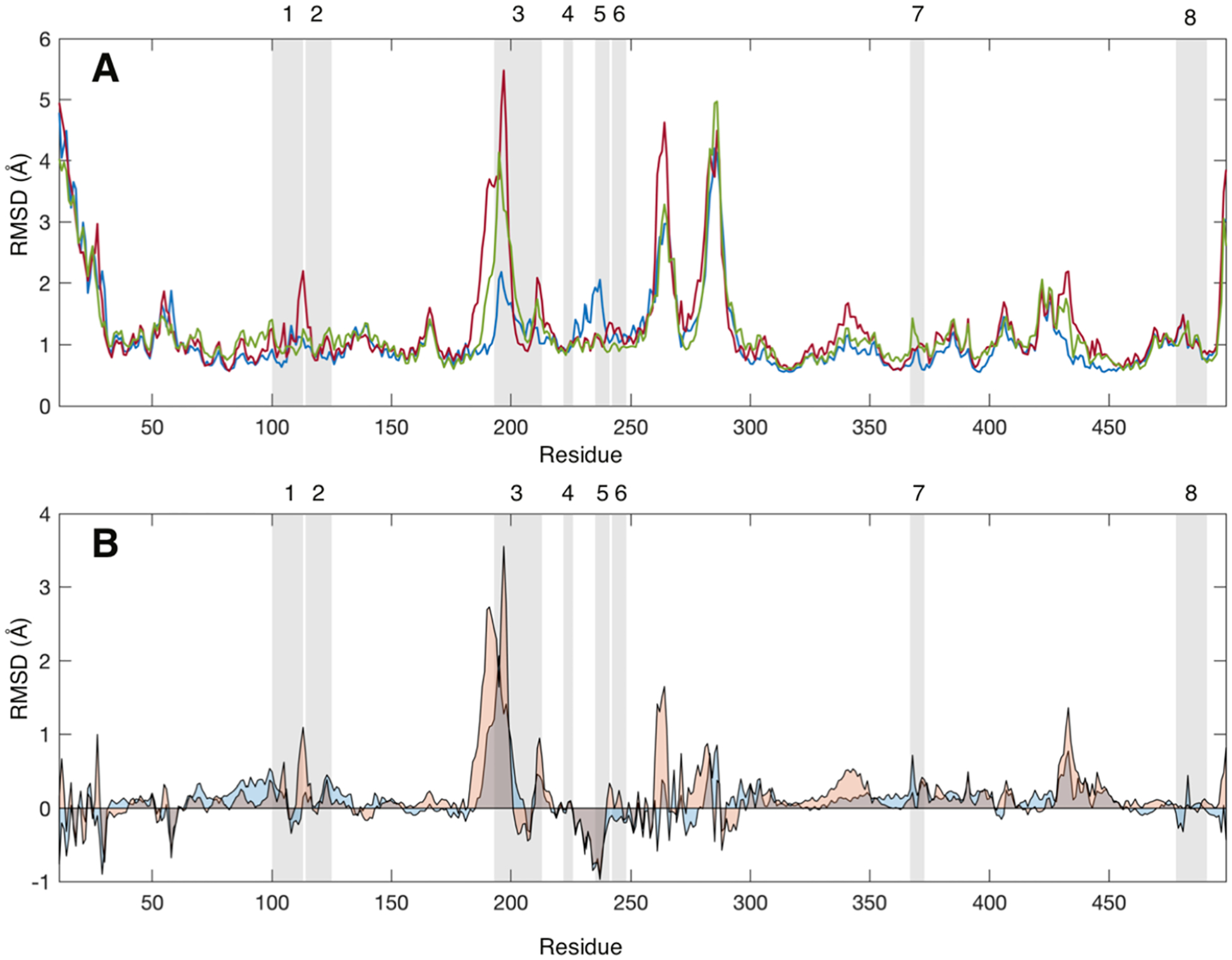
(A) Average root-mean-square deviation (RMSD, Å) values for each CYP3A4 residue without a ligand (blue), with one MDZ (red), and with two MDZ (green) bound in the active site. (B) Average RMSD values for each CYP3A4 with one MDZ (blue) or two MDZ (orange) less the average values for the ligand-free enzyme. RMSD values were calculated relative to the average structure over the entire trajectory. Gray-shaded regions highlight the peptides identified in the mass spectrometry experiments. RMSD values were calculated using the CA, CB C, O, N, HA, HA1, HA2, and HN atoms for all frames in the latter 0.4 μs of each simulation.
