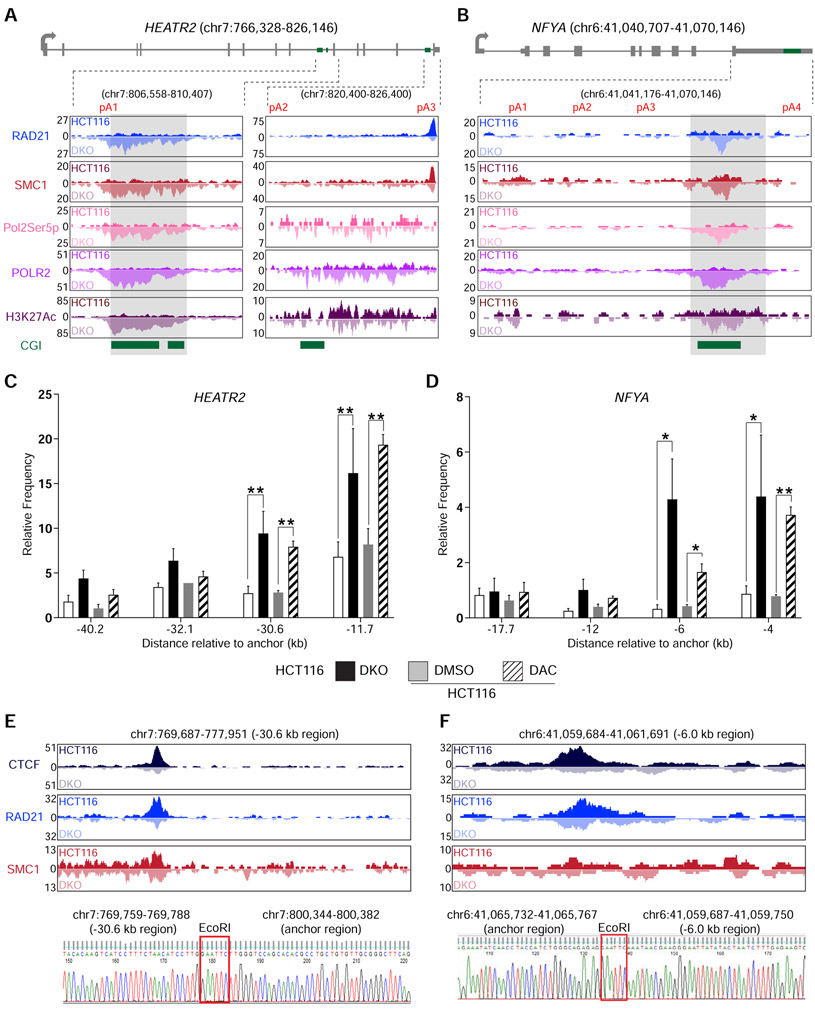
Figure 3: DNA methylation-sensitive binding of CTCF and the cohesin complex orchestrate intra-chromosomal looping.
a, b, Gene model and UCSC genome browser views for HEATR2 (a) and NFYA (b). ChIP-seq signals for RAD21, SMC1, RNA Polymerase II phosphorylated at serine 5 (Pol2Ser5p), Total RNA polymerase II (POLR2), and histone H3 acetylated lysine 27 (H3K27Ac) are shown. CGI, annotated CpG islands. c, d, 3c-qPCR analysis of long-distance intra-chromosomal loops at HEATR2 (c) and NFYA (d). Relative contact frequencies between the anchor fragment and distal fragments were normalized to control BAC clones. n = 3 biological replicates. Data are represented as mean ± SEM. *, p <0.05; **, p <0.005. e, f, Visualization and confirmation of 3C-detected contacting genomic fragments for HEATR2 (e) and NFYA (f). Top, UCSC genome browser views of the −30.6 kb region for HEATR2 (e) and the −6.0 kb region for NFYA (f). ChIP-seq signals for CTCF, RAD21, and SMC1 are shown for each fragment. Bottom, Sanger sequencing results showing the ligation between the anchor regions and the distal contact regions in HEATR2 and NFYA. EcoRI, EcoRI restriction enzyme site used in the 3C experiment. See also Figure S3 and Table S4.