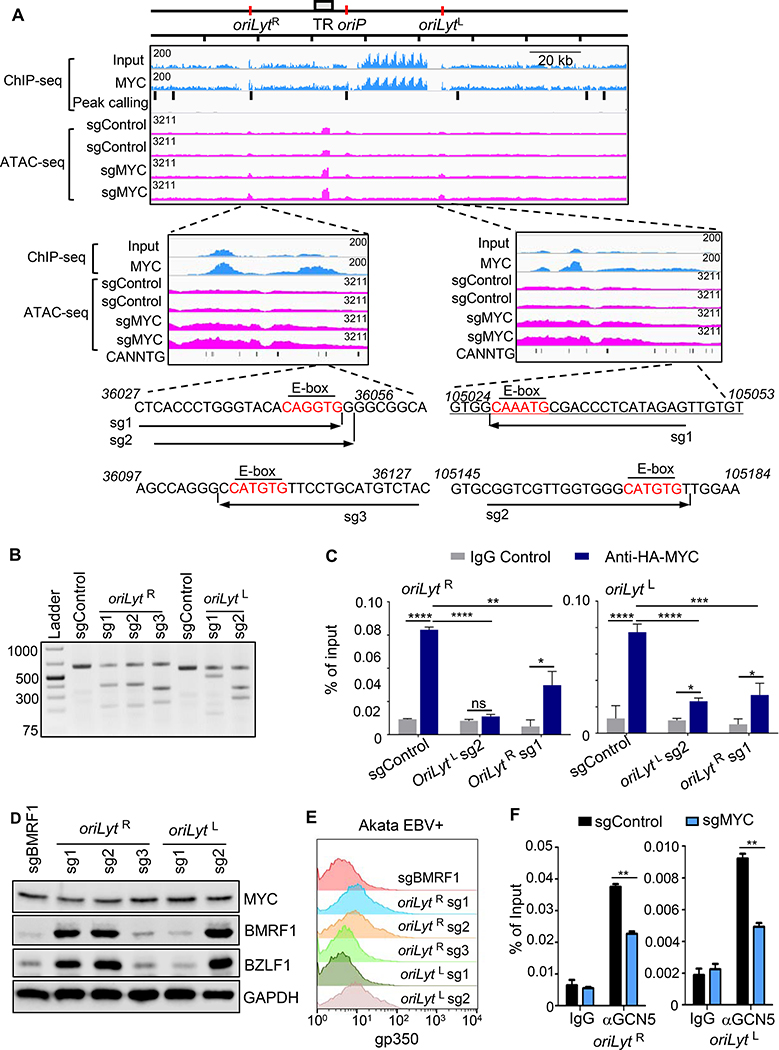
Figure 6. MYC Occupancy of EBV Genome oriLyt E-Box Sites Maintains Latency in BL.
(A) ChIP-seq analysis of EBV genome MYC occupancy and ATAC-seq analysis of effects of MYC depletion on EBV genomic accessibility. Shown are Daudi BL EBV genome input and MYC ChIP-seq tracks. Background-subtracted peak calling identified seven significant EBV genomic peaks, highlighted by black bars. Values at top left indicate track heights. n=2 ATAC-seq tracks from Akata with indicated sgRNAs and treated with acyclovir (100 μg/ml). Zoomed tracks of oriLyt regions are shown. Akata EBV DNA sequences of sgRNA-targeted regions are shown, with E-boxes in red.
(B) T7E1 assay of Cas9 oriLyt region editing. Representative T7E1 nuclease-treated PCR products are shown.
(C) ChIP-qPCR of MYC oriLyt region occupancy. Ig-control or anti-HA ChIP was performed on chromatin from Akata with HA-MYC and indicated sgRNA expression. qPCR was done with E-box regions primers. Mean + SD are shown for n=3 replicates. ****p < 0.0001, **p < 0.001, **p<0.01.
(D) Immunoblot of WCL from Akata with indicated sgRNAs. Blot is representative of n=3.
(E) FACS PM gp350 abundances in Akata with indicated sgRNAs.
(F) ChIP-qPCR analysis of STAGA subunit GCN5 occupancy at oriLyt regions. αGCN5 ChIP was performed on chromatin from Akata with control or MYC sgRNAs, followed by qPCR using primers specific for oriLyt E-box regions. Mean + SD are shown for n=3 replicates. **p<0.01.
See also Figure S7.