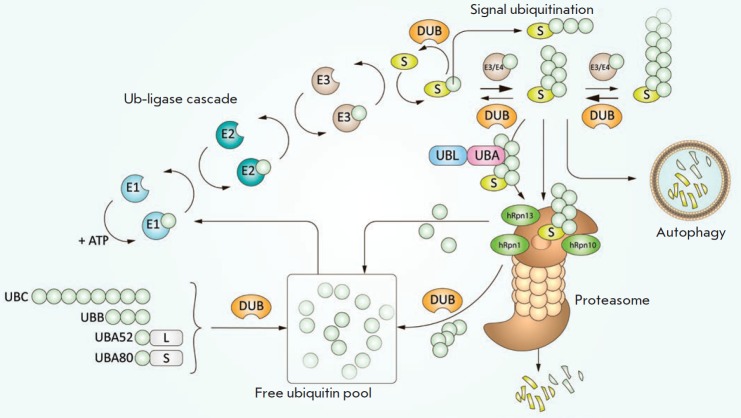
Fig. 1.
Ubiquitin-proteasome system. Ubiquitin is synthesized in the form of four precursor proteins, UBC, UBB, UBA52, and UBA80, which are further processed by specialized deubiquitinating enzymes (DUBs)–ubiquitin-isopeptidases. The ubiquitination system, which includes three types of ubiquitin ligases (E1 (two enzymes), E2 (tens of enzymes), and E3 (hundreds of enzymes)), is highly specific and selective due to its hierarchical structure. Ubiquitin is conjugated to a substrate (S) as a monomer or a polyubiquitin chain that is formed through the internal lysine residues of the preceding Ub. The polyubiquitin chain is elongated by E3 ligases or the relatively recently discovered ubiquitin ligases of the E4 family. There is a dynamic equilibrium between ubiquitination and removal of ubiquitin residues by ubiquitin isopeptidases, which controls the optimal chain length that amounts, according to modern concepts, to about six ubiquitin molecules per substrate molecule [8]. Further, the ubiquitinated substrate binds to the Rpn10, Rpn13, and Rpn1 proteasome subunits, either directly or with the participation of shuttle proteins of the UBL–UBA family. There may be specific autophagy. A certain amount of ubiquitin enters the proteolytic chamber together with the substrate, which leads to its degradation. In most cases, proteasome deubiquitinase Rpn11 successfully removes the entire polyubiquitin chain that is further cleaved into monomers for recycling