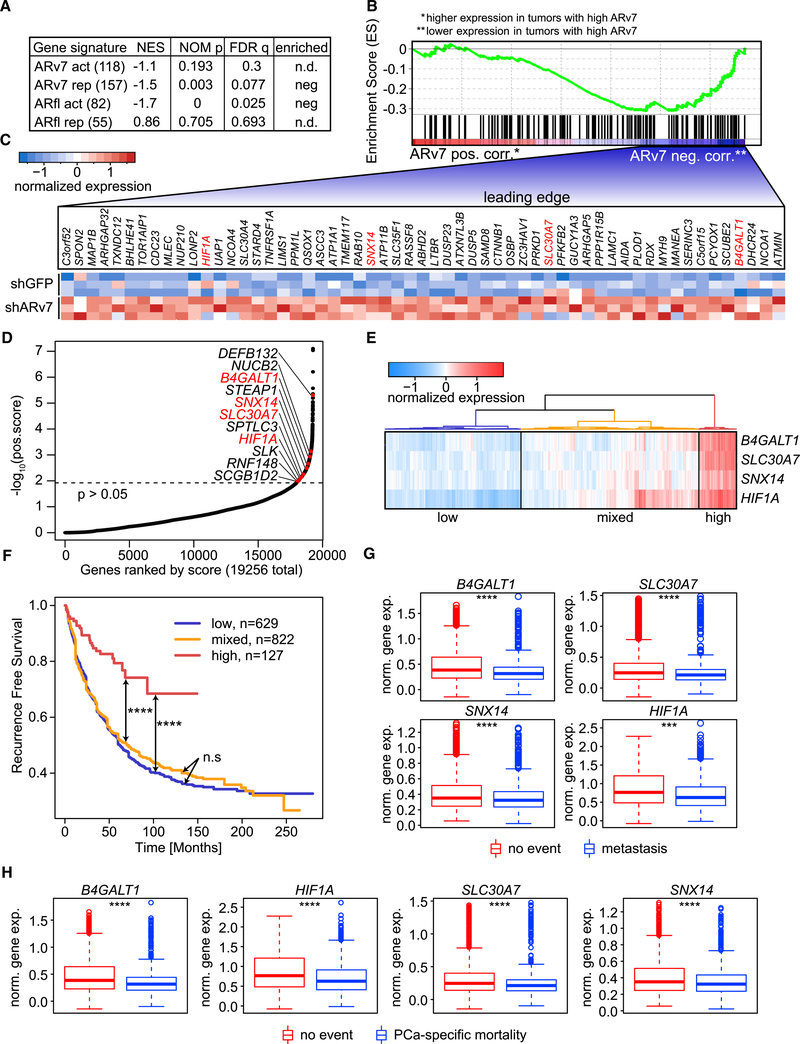
Figure 6. ARv7 Represses Genes with a Tumor-Suppressive Function.
(A) GSEA of ARfl- and ARv7-specific gene signatures (Table S5), compared with genes ranked by transcriptome data from CRPC tumors with IHC-defined ARv7 expression. The number of genes in each signature is indicated in parentheses. The normalized enrichment score (NES), the nominal p value (NOM p), and the FDR q value (FDR q) are shown.
(B) GSEA-determined enrichment profile of the ARv7-repressed gene signature (ARv7 rep) as shown in (A). Genes are ranked by their expression in IHC-defined ARv7 high (red, left, ARv7 pos. corr.) versus low patient tumors (blue, right, ARv7 neg. corr.).
(C) Heatmap of relative expression of 57 target genes defined by the leading edge in (B), in shGFP and shARv7 LNCaP95 cells. Genes in red were also identified in (D).
(D) Hockey-stick plot of positively selected genes identified in a genome-wide CRISPR KO screen. Genes were ranked according to their r score and log10 p value. Target genes from the ARv7-repressed gene signature in (A) are indicated, and genes highlighted in red overlap with the leading-edge analysis in (C).
(E) Hierarchical clustering of the 4 ARv7-target genes from (D) on gene expression data from patient samples (Decipher-GRID). Three clusters, based on the average expression of the four genes, are shown: low (blue), mixed (orange), and high (red).
(F) Kaplan-Meier graphs of prostate-specific antigen recurrence-free survival for the three patient clusters defined in (E). Log-rank test: n.s, not significant; ****p < 0.0001.
(G and H) Expression of the 4 ARv7-regulated genes in (D) in patient subgroups (Decipher-GRID). Samples are grouped by patients that did (n = 492) or did not (n = 1,134) develop metastasis (G), or did (n = 236) or did not (n = 1,211) succumb to the tumor (PCa-specific mortality; H). Standard t test: ***p % 0.001, ****p % 0.0001. Box plots show the median, and the first and third quartile. Whiskers extend to 1.5 the interquartile range and data beyond that are shown as individual points.