Figure 1.
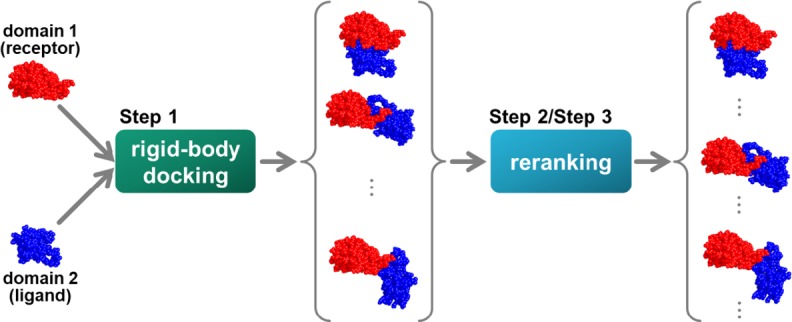
Problem setting. Step 1: We obtained a two-domain protein from the PDB and divided each domain according to the definition. Next, we generated a structural model using a rigid-body docking tool with two input domain structures. Step 2: We calculated some scores for each structural model. Step 3: We evaluated and reranked each model with a score function using some scores. As a result, the prediction was considered as successful when there was at least one acceptable structure within the top 10 solutions.
