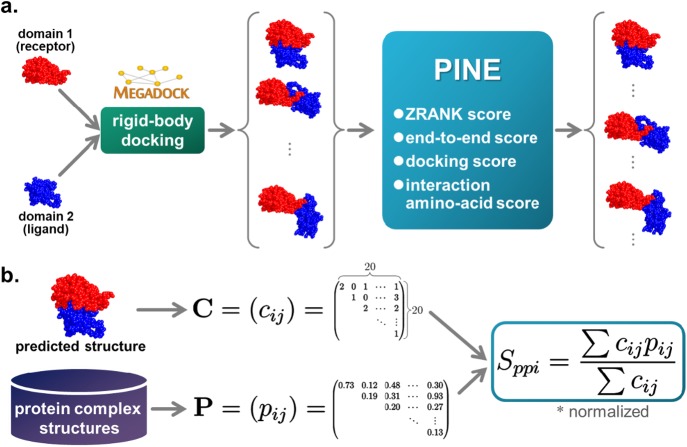
Figure 2.
a. Outline of the proposed method. Step 1: Obtain domains as in the existing method and generate 10,800 structural models for each multidomain protein with the rigid-body docking tool MEGADOCK. Docking score is calculated at the time of this docking calculation. Step 2: Calculate the proposed score for each structural model. Step 3: Rerank the structural model with the score function PINE using each score and predict the structure. b. Calculation of the interaction amino acid residue score Sppi. The PPI matrix P was generated from protein complex structures in advance and the domain contact matrix C was generated from a predicted two-domain protein structure. Next, Sppi of the predicted structure was calculated with P and C. This score predicts interactions without requiring a template for the overall structure of the multidomain protein.