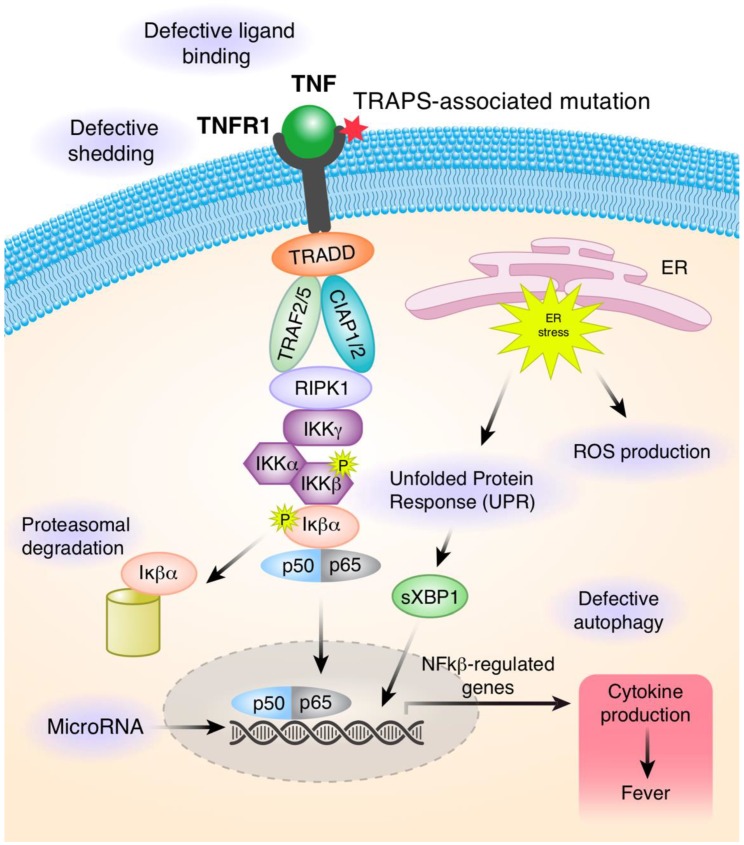
Figure 2.
Summary of proposed pathogenic mechanisms in TRAPS. Binding of TNFα to TNFR1 leads to the assembly of the signaling pathway that ultimately upregulates the gene expression of many pro-inflammatory cytokines. There are multiple mechanisms that contribute to the pathogenesis of TRAPS. Heterozygous variants affect the structure of the extracellular domain and impact its ability to bind to the TNF ligand. Mutant receptors fail to shed from the cell surface to generate soluble TNFR1 proteins, which function to attenuate signaling through the TNFR1 receptor. Mutated misfolded proteins accumulate in the cells and cause endoplasmic stress (ER), upregulation in the unfolded protein response (UPR), and increased production of mitochondrial reactive oxygen species (ROS). The UPR initiates ER membrane stress sensors, including inositol-requiring protein (IRE1)α, to restore protein folding and homeostasis in the ER. In the ER stress, activation of IRE1α leads to splicing of transcription factor X-box binding protein 1 (XBP1) into its active form sXBP1, which acts as a transcription factor that can upregulate expression of many target genes. Autophagy is responsible for clearance of intracellular TNFR1. However, in patients with TRAPS, autophagy is defective and mutated proteins are not efficiently cleared from cells. MicroRNA can regulate gene expression at the transcriptional and post-transcriptional levels by binding to the complementary mRNA sequence. MicroRNAs can be detected in serum and various miRNAs can serve as biomarkers of the disease activity.