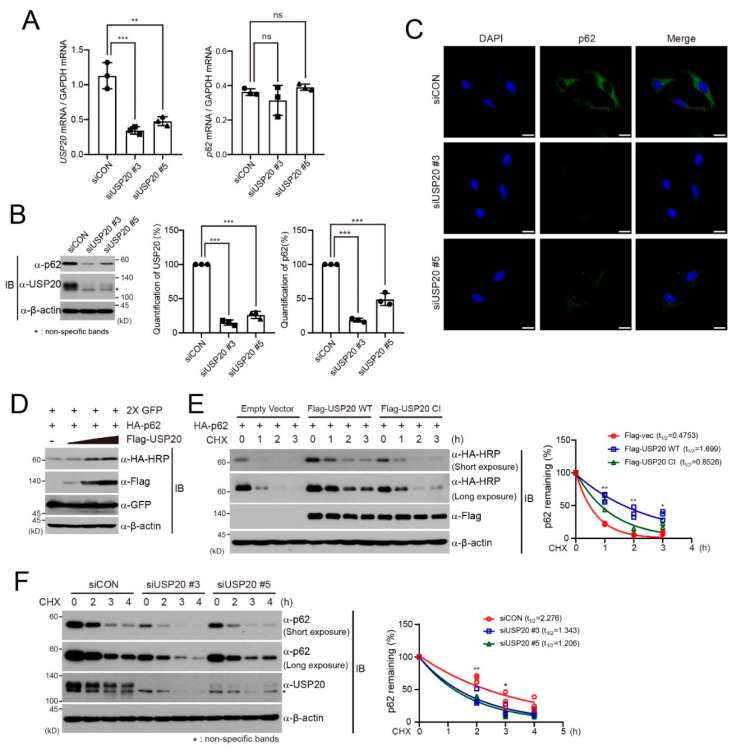
Figure 1.
USP20 is required for p62 protein stability. (A) HeLa cells were reverse-transfected with 20-nM control siRNA (siCON) or one of two independent USP20-specific siRNAs (siUSP20 #3 and siUSP20#5). USP20 and p62 mRNA levels were measured by quantitative real-time reverse -transcriptase-polymerase chain reaction (qRT-PCR) in control and USP20-depleted HeLa cells and normalized to Gapdh mRNA. (B) Total cell lysates obtained from USP20-depleted and control HeLa cells were immunoblotted with the indicated antibodies. Expression levels of endogenous p62 and USP20 proteins were quantified by using ImageJ software. For normalization, β-actin expression was used as a control. (C) Endogenous expression of p62 protein in USP20-depleted and control HeLa cells were detected by immunofluorescence analysis. Scale bars, 20 µm. (D) After a plasmid encoding HA-p62 was co-transfected into HEK293 cells with dose-dependent expression of Flag-USP20, cells were immunoblotted with the indicated antibodies. Expressions of green fluorescence protein (GFP) and β-actin were used as loading controls. (E) After plasmids encoding wild-type (WT) Flag-USP20 or a catalytically inactive (CI) mutant of Flag-USP20 were respectively transfected into HeLa cells, cells were treated with 50 μg/mL cycloheximide (CHX) for the indicated times and lysates were immunoblotted with the indicated antibodies. Empty vector was used as a control. (F) USP20-depleted or control (siCON) HeLa cells were treated with 50 μg/mL CHX for the indicated times. Total cell lysates were immunoblotted with the indicated antibodies. In (E) and (F), p62 levels were quantified by ImageJ software and normalized to β-actin expression. Data were statistically analyzed by two-way ANOVA followed by Bonferroni’s multiple comparison test (* p < 0.05, ** p < 0.01, *** p < 0.001 compared to siCon or empty vector, ns; not significant, n = 3). Bars represent the mean ± SD. Images are representative of three independent experiments.