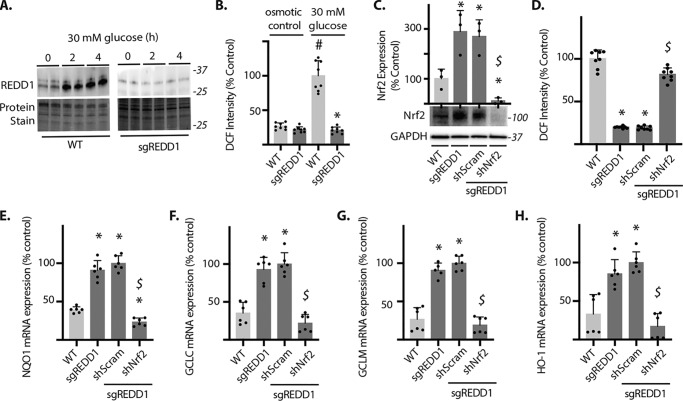
Figure 2.
Nrf2 is necessary for the attenuation of oxidative stress in REDD1-deficient cells exposed to hyperglycemic conditions. A, WT and REDD1 CRISPR knockout (sgREDD1) MIO-M1 cells were cultured in medium containing 5 mm glucose and exposed to culture medium containing 30 mm glucose. REDD1 protein expression was evaluated in cell lysates by Western blotting. Protein molecular mass (in kilodaltons) is indicated at the right of blots. Protein loading was evaluated by reversible protein stain. B, WT and sgREDD1 cells were exposed to medium containing either 30 mm glucose or 5 mm glucose plus 25 mm mannitol (osmotic control) for 4 h. ROS were visualized with DCFDA and DCF fluorescent intensity was quantified. C-H, Nrf2 was knocked down by stable expression of shRNA (shNrf2). Data in C–H were obtained using sgREDD1 Nrf2 knockdown clone 1 from Fig. S1. Control sgREDD1 cells expressed a scramble shRNA (shScram). All cells in C–H were exposed to medium containing 30 mm glucose for 4 h prior to analysis. C, Western blotting was used to evaluate Nrf2 and GAPDH protein expression in whole cell lysates. D, DCFDA was used to visualize ROS and DCF intensity was quantified. The abundances of mRNAs encoding NQO1, GCLC, GCLM, and HO-1 were evaluated in cell lysates by PCR in E-H, respectively. Values are mean ± S.D. #, p < 0.05 versus osmotic control. *, p < 0.05 versus WT. $, p < 0.05 versus shScram.