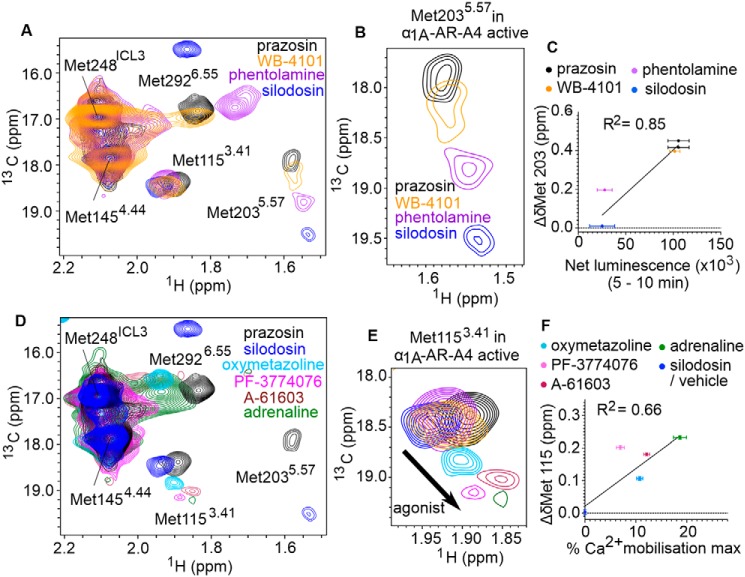
Figure 5.
1H-13C SOFAST-HMQC spectra of α1A-AR-A4–active. A, Overlay of 2D 1H-13C SOFAST-HMQC spectra of ([13CϵH3]Met-) α1A-AR-A4–active bound to prazosin (black, inverse agonist), WB-4101 (orange, inverse agonist), phentolamine (purple, inverse agonist), and silodosin (blue, neutral antagonist). B, close-up of the Met-2035.57 resonance. C, linear regression analysis of the average chemical shift differences (Δδ) for the 13CϵH3 of Met-2035.57 in α1A-AR-A4–active when bound to prazosin (black circles), WB-4101 (orange circles), and phentolamine (purple circles) compared with silodosin (blue circles) and the increase in luminescence seen in the NanoBit assay with α1A-AR-A4–active expressing COS-7 cells treated with the same antagonist (from Fig. S11A). Testing the resultant equation against the null hypothesis of a slope of zero resulted in a p value of 0.0041. D, overlay of 2D 1H-13C SOFAST-HMQC spectra of ([13CϵH3]Met-) α1A-AR-A4–active bound to prazosin (black, inverse agonist), silodosin (blue, neutral antagonist), oxymetazoline (cyan, partial agonist), PF-3774076 (magenta, partial agonist), A-61603 (maroon, full agonist), and adrenaline (green, full agonist). E, close-up of the Met-1153.41 resonance. F, linear regression analysis of the average chemical shift differences (Δδ) for the 13CϵH3 of Met-1153.41 in α1A-AR-A4–active when bound to oxymetazoline (cyan circles), PF-3774076 (pink circles), A-61603 (dark red circles), adrenaline (green circles), and silodosin (blue circles) and the efficacy of each agonist in triggering Ca2+ mobilization in α1A-AR-A4–active expressing COS-7 cells (from Fig. S8, B–E). Testing the resultant equation against the null hypothesis of a slope of zero resulted in a p value of 0.0154. C and F, Δδ are plotted for two independent titrations of prazosin, silodosin, and oxymetazoline, and single experiments for other ligands. In A, B, D, and E, spectra were acquired on ∼50 μm α1A-AR-A4–active dissolved in 0.02–0.1% DDM micelle, pH 7.5, and 25 °C. Average chemical shift differences (Δδ) were normalized using the equation Δδ = [(Δδ1H)2 + (Δδ13C/3.5)2]0.5 and errors were calculated by the formula |[Δδ1H × R1H + Δδ13C × R13C/(3.5)2]|/Δδ, where R1H and R13C are the digital resolutions in ppm in the 1H and 13C dimensions respectively (5).