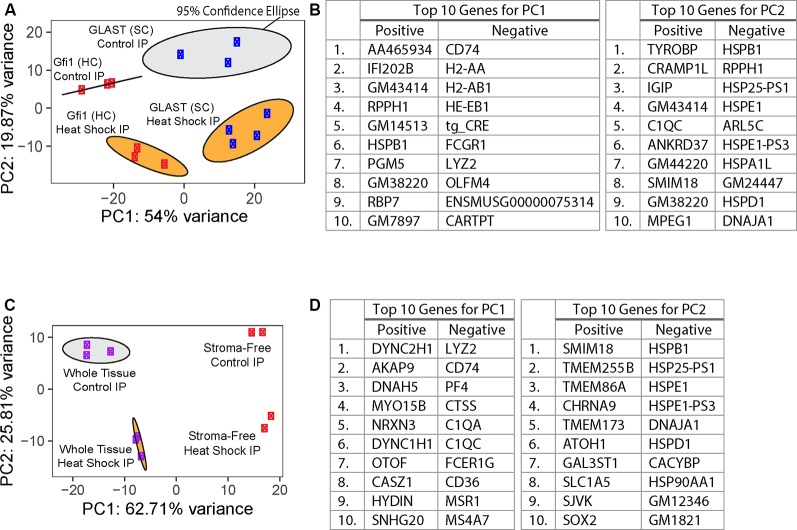
Figure 3.
Principal component analysis (PCA) of RiboTag IP samples. (A) PCA of whole-tissue IP samples from RiboTag Gfi1-Cre (red) and GLAST-CreER (blue) IPs. PC1 represents 54% of the total variance in the experimental data, and PC2 represents 19.87% of the total variance. Ellipses represent 95% confidence ellipses around each group of samples, and the color of each ellipse corresponds to the treatment type (heat shock in orange or control in gray). (B) Tables show the 10 genes with the highest loadings in either direction for the two PCs plotted in (A). (C) PCA of IP samples from RiboTag Gfi1-Cre whole-tissue (purple) and stroma-free (red) IPs. PC1 represents 62.71% of the total variance in the experimental data, and PC2 represents 25.81% of the total variance. Ellipses represent 95% confidence ellipses around each group of at least three samples, and the color of each ellipse corresponds to the treatment type (heat shock in orange or control in gray). (D) Tables show the 10 genes with the highest loadings in either direction for the two PCs plotted in (C).