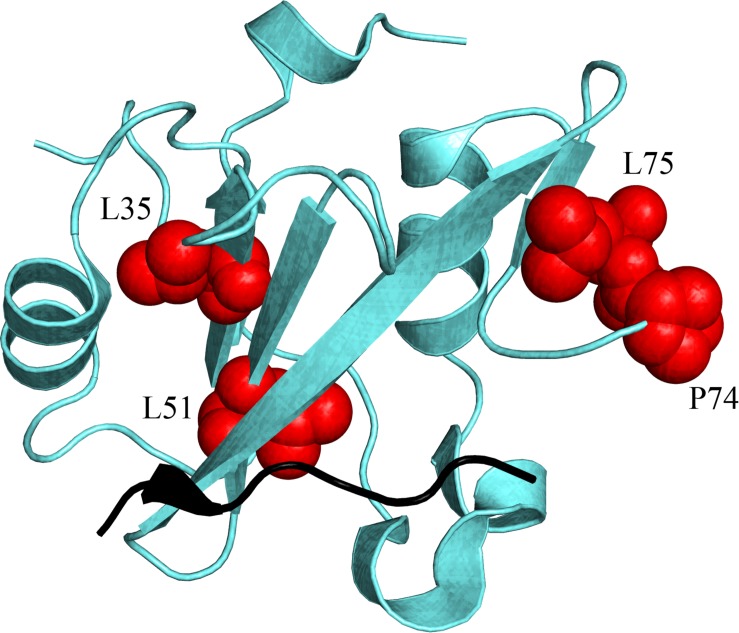
FIGURE 5.
Structural distribution of the residues (highlighted in red on N-SH2 structure) that are energetically coupled with M residue part of the pY-X-X-M consensus that is specifically recognized by N-SH2, and finely modulate the affinity of the domain for its natural binding partners (see text for details). Because of the unavailability of the N-SH2:Gab2 complex structure, a different ligand is represented in black (cKit) only to pinpoint the position of the binding pocket of N-SH2. (PDB: 2IUH).